factorMerger
Overview
factorMerger is a set of tools to support post-hoc testing that would enable to extract hierarchical structure of factors with respect to a given response.
Installation
# the easiest way to get factorMerger is to install it from CRAN:
install.packages("factorMerger")
# Or the the development version from GitHub:
# install.packages("devtools")
devtools::install_github("ModelOriented/factorMerger", build_vignettes = FALSE)More
The description of factorMerger may be found in the paper The Merging Path Plot: adaptive fusing of k-groups with likelihood-based model selection (here).
Workflow
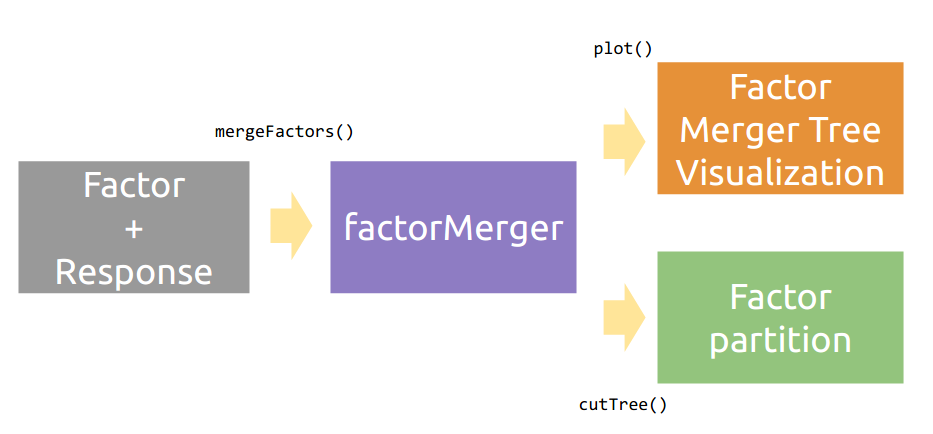
Cheatsheet
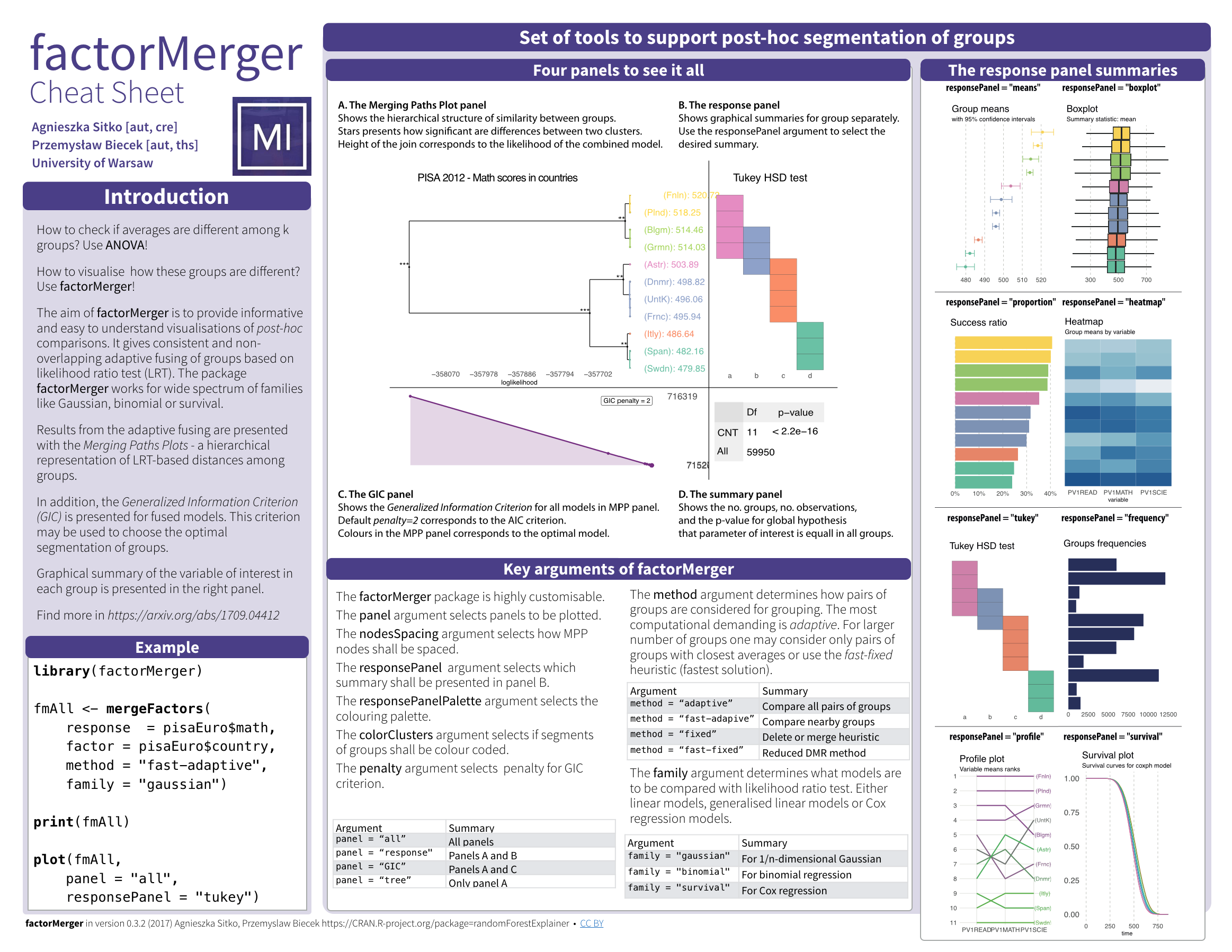
Example
Survival analysis
library(factorMerger)
library(forcats) # distinguish meaningful factors (fct_lump)
data(BRCA)
brcaSurv <- survival::Surv(time = BRCA$time, event = BRCA$vitalStatus)
drugName <- fct_lump(as.factor(BRCA$drugName), prop = 0.05)
drugNameFM <- mergeFactors(response = brcaSurv[!is.na(drugName)],
factor = drugName[!is.na(drugName)],
family = "survival")
plot(drugNameFM, nodesSpacing = "effects", gicPanelColor = "grey2")
