One option is to use VIVO/GRID: https://grid.ac/downloads
Not clear if this best. It's large, not clear how easy to add, and the entities we care about are not just institutions. We also want to give provenance to specific projects (typically funded).
We will most likely maintain our own list of organizations as a yaml that can be easily updated.
-
id: https://www.ucl.ac.uk/functional-gene-annotation/cardiovascular
label: Cardiovascular Gene Annotation
-
...with the obvious json-ls/ttl translation. We can add owl:sameAs for GRID IDs if required.
@mellybelly thoughts?
We will use these using our ugly entity-as-literal hack for now (cc @balhoff ), so the ttl would be:
dc:??? "https://www.ucl.ac.uk/functional-gene-annotation/cardiovascular"^^xsd:String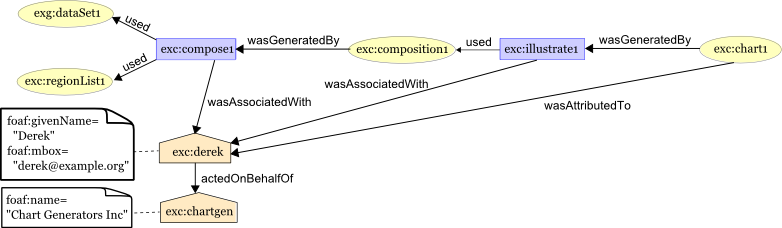
This is that "hats" concept that floats up occasionally (that I thought was already captured somewhere, but I have been unable to find).
While the ORCID-per-annotation model gets us a long way, individuals work for different entities over time, and at the same time, so there needs to be some method not only capturing who did something, we also have to capture what "hat" they were wearing when they did it.
The noctua bit of this, besides proper message passing, would be a dropdown of available hats for the logged-in user.
Somewhat related to #347, as the same kind of group information is a prerequisite. The group/funding information would likely need to be partially kept in users.yaml, with maybe an ontology or reference IDs for group/funding entities (TBD, and maybe an item for geneontology/go-site).