Softcite dataset
A gold-standard dataset of software mentions in research publications for supervised learning based named entity recognition, by Howison Lab at the University of Texas at Austin and Science-miner, with funding support from the Sloan Foundation.
This repository is for v1 of the dataset. We have since published v2 with additional data and annotation codes: https://github.com/softcite/softcite_dataset_v2 and https://zenodo.org/record/7995565.
Why creating this dataset
Software lays a critical foundation for measureless research activities today. However, researchers are often frustrated by redundant, incompatible, or poorly supported pieces of software (Howison et al., 2015). One pathway to improve software for research is to increase the visibility of software in the bibliometric-based system of research impact. So that software contributions to research can be well acknowledged, software creators, funders, and other stakeholders have more incentives to cooperate and provide more sound, quality-assured software work.
We have annotated ~5k open access research publications in areas of life sciences and social sciences. As a result, we identified 4,093 software mentions in these publications. Largely these software mentions are not indexed, formal citations (Howison & Bullard, 2016). So we have ferreted out a lot of software that contributed to research but currently are not visible to academic databases and systems of information retrieval.
Dataset content
Our released dataset is a TEI/XML corpus file that contains metadata of annotated research publications and software mentions identified in these publications. The software mentions are further annotated with details about the software, including software version, publisher, and access url, if present in the publication text. The annotated software mentions are presented in their paragraph context considering that they are contextual specific. Below is an example of software mentions in annotated publication with all metadata encoded with the TEI/XML schema. You can find thousands of entries like this in the Softcite dataset.
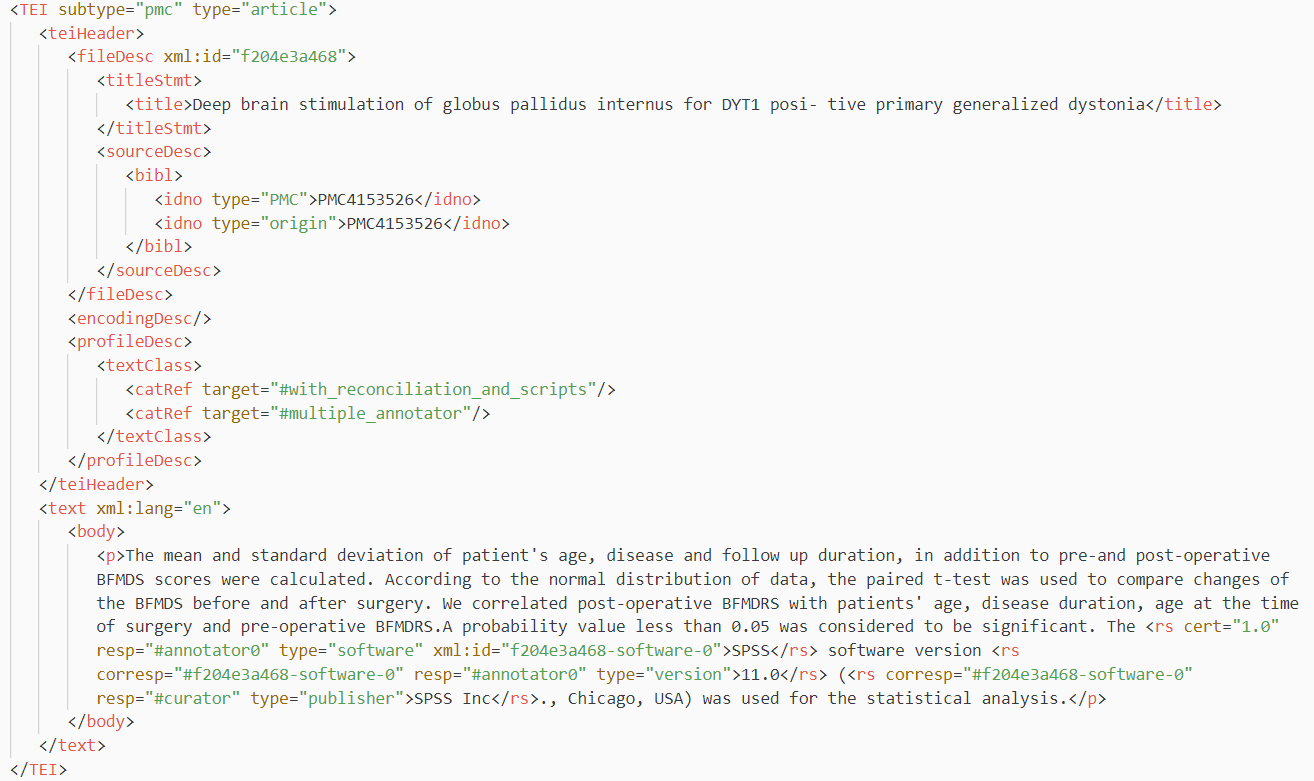 Image: Snapshot of an annotated article entry with encoded software annotations in the Softcite dataset
Image: Snapshot of an annotated article entry with encoded software annotations in the Softcite dataset
Use scenarios
We created the Softcite dataset in a machine-readable (TEI/XML) format for immediate machine learning use. It is designed to accommodate training/validation for supervised learning based scholarly text mining. You could use it to train your model for software entity recognition in text, to develop utilities for increasing software visibility to information systems, or to investigate how software has been used for research. We have prototyped machine learning training ourselves and validated that the Softcite dataset is effective for machine learning use. If you need help for a safe jumpstart for your project, feel free to create a Discussion (or email Fan Du (@caifand) if for some reason discussion on a public forum is not possible).
The Softcite approach
Understanding data provenance is important for data reuse. We have a forthcoming publication that details the design consideration and creation process of the Softcite dataset. It also documents the annotation schema of the Softcite dataset.
To ensure data consistency across the whole pipeline, we used GROBID, an open source machine learning library, to convert thousands of open access publication PDFs into TEI/XML text. Our annotation team consists of experts in NLP and research software, and students from UT Austin who had been through group training. (Training materials can be found here) The annotation team read through every PDF publication, annotated mentions of software and their details. We conducted an inter-annotator agreement check to ensure training was effective. We collected all the annotation data input from annotators via GitHub and validated the incoming data via a test suite deployed on Travis CI, coupled with senior annotators' manual examination.
 Image: Collective annotation workflow for Softcite dataset creation
Image: Collective annotation workflow for Softcite dataset creation
After the annotation, we checked the annotation consistency across the whole dataset via a script and expert review. All the software mentions in the Softcite dataset have been reviewed by expert annotators, ensuring its gold standard quality. We encoded the annotations in the converted TEI/XML article text using GROBID.
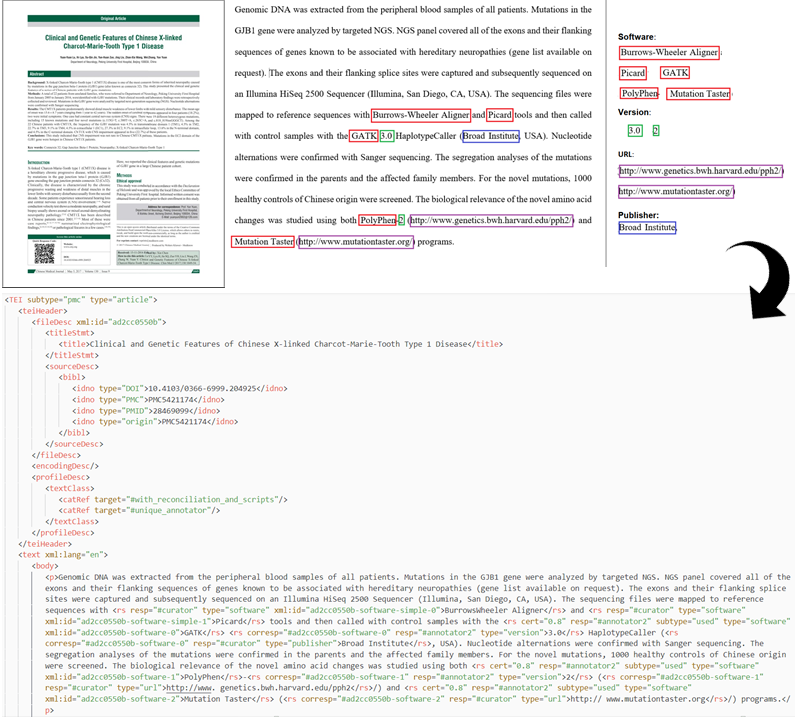 Image: From annotating software mentions in PDFs to a TEI/XML corpus
Image: From annotating software mentions in PDFs to a TEI/XML corpus
Release and Change
The first formal and latest release is softcite-dataset v1.0. It is also available in Zenodo. For changes to the dataset, please see our changelog.
Implementation
We have utilized the Softcite dataset to train a set of machine learning models and implemented them in a GROBID module for software mention recognition. The software mention recognizer is further used to seed a Software Knowledge Base. The Software Knowledge Base provides capabilities such as software entity extraction, disambiguation, citation information retrieval, and cross-entity inference. A REST API is provided for leveraging the Software Knowledge Base capabilities.
Roadmap
- We hope to build interoperability with other existing datasets and resources about software mentions in scientific text. Interoperable datasets can scale up ML-based efforts for software entity recognition and increasing research software visibility.
- We also have developed CiteAs.org in collaboration with Our Research. CiteAs.org is an interactive search engine for discovering software and other research outputs online. It offers citation recommendation and provenance according to your search query, and our goal is to integrate the Software Knowledge Base into CiteAs.org for enhanced recommendation.
- We have crowdsourced more software annotation data based on our annotation scheme in different domain literature. We expect to release this data for the community working on software entity recognition in the future.
If you have any suggestions, comments, welcome to start an issue or discussion in this repository, or contact Fan Du (@caifand.
License

The Softcite dataset is licensed under a Creative Commons Attribution 4.0 International License.
Acknowledgement
We thank Alfred P. Sloan Foundation for supporting this work. We also appreciate our collaborators and student annotators for making this dataset gold-standard and available.
References
Howison, J., Deelman, E., McLennan, M. J., Ferreira da Silva, R., & Herbsleb, J. D. (2015). Understanding the scientific software ecosystem and its impact: Current and future measures. Research Evaluation, 24(4), 454-470. DOI: 10.1093/reseval/rvv014
Howison, J., & Bullard, J. (2016). Software in the scientific literature: Problems with seeing, finding, and using software mentioned in the biology literature. Journal of the Association for Information Science and Technology, 67(9), 2137-2155. DOI: 10.1002/asi.23538
Du, C., Cohoon, J., Lopez, P., & Howison, J. (forthcoming). Softcite Dataset: A Dataset of Software Mentions in Biomedical and Economic Research Publications. Journal of the Association for Information Science and Technology. DOI: 10.1002/asi.24454