obistools: Tools for data enhancement and quality control.
Installation
Taxon matching
Check required fields
Plot points on a map
Identify points on a map
Check points on land
Check depth
Check eventID and parentEventID
Check eventID in an extension
Flatten event records
Flatten occurrence and event records
Calculate centroid and radius for WKT geometries
Map column names to Darwin Core terms
Check eventDate
Data quality report
Lookup XY
Installation
Installing obistools requires the devtools package:
install.packages("devtools")
devtools::install_github("iobis/obistools")Taxon matching
match_taxa() performs interactive taxon matching with the World Register of Marine Species.
names <- c("Abra alva", "Buccinum fusiforme", "Buccinum fusiforme", "Buccinum fusiforme", "hlqsdkf")
match_taxa(names)3 names, 1 without matches, 1 with multiple matches
Proceed to resolve names (y/n/p)? y
AphiaID scientificname authority status match_type
1 531014 Buccinum fusiforme Kiener, 1834 unaccepted exact
2 510389 Buccinum fusiforme Broderip, 1830 unaccepted exact
Multiple matches, pick a number or leave empty to skip: 2
scientificName scientificNameID match_type
1 Abra alba urn:lsid:marinespecies.org:taxname:141433 near_1
2 Buccinum fusiforme urn:lsid:marinespecies.org:taxname:510389 exact
2.1 Buccinum fusiforme urn:lsid:marinespecies.org:taxname:510389 exact
2.2 Buccinum fusiforme urn:lsid:marinespecies.org:taxname:510389 exact
3 <NA> <NA> <NA>Check required fields
check_fields() will check if all OBIS required fields are present in an occurrence table and if any values are missing.
data <- data.frame(
occurrenceID = c("1", "2", "3"),
scientificName = c("Abra alba", NA, ""),
locality = c("North Sea", "English Channel", "Flemish Banks"),
minimumDepthInMeters = c("10", "", "5")
)
check_fields(data)This function returns a dataframe of errors (if any):
field level message row
1 eventDate error Required field eventDate is missing NA
2 decimalLongitude error Required field decimalLongitude is missing NA
3 decimalLatitude error Required field decimalLatitude is missing NA
4 scientificNameID error Required field scientificNameID is missing NA
5 occurrenceStatus error Required field occurrenceStatus is missing NA
6 basisOfRecord error Required field basisOfRecord is missing NA
7 scientificName error Empty value for required field scientificName 2
8 scientificName error Empty value for required field scientificName 3Plot points on a map
plot_map() will generate a ggplot2 map of occurrence records, plot_map_leaflet() creates a Leaflet map.
plot_map(abra, zoom = TRUE)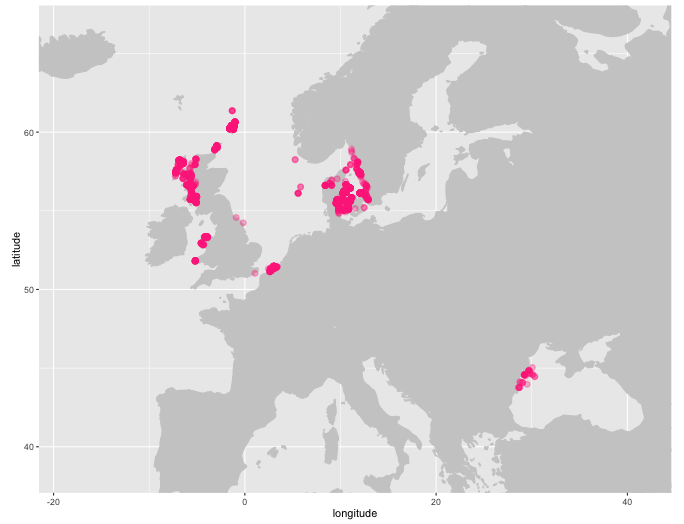
plot_map_leaflet(abra)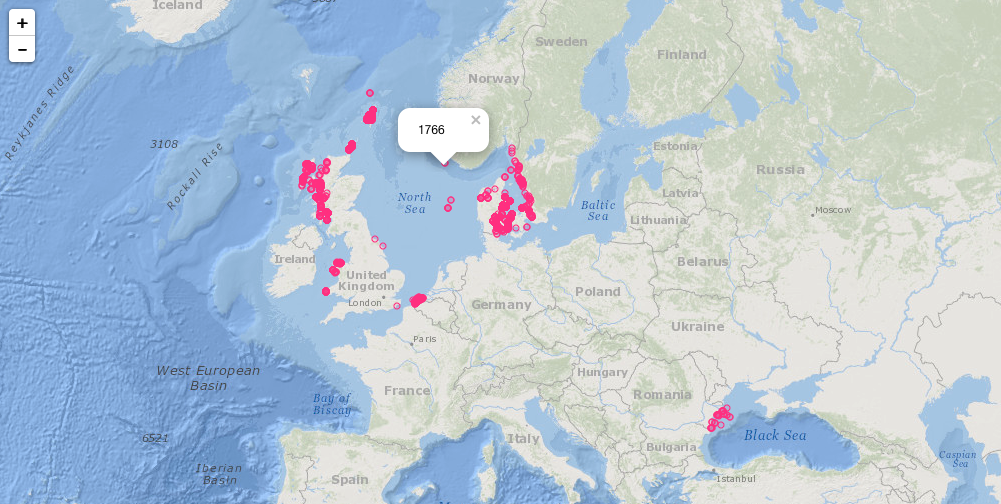
Identify points on a map
Use identify_map() to identify points on a ggplot2 map. This function will return the record closest to where the mouse was clicked.
plot_map(abra, zoom = TRUE)
identify_map(abra) id decimalLongitude decimalLatitude basisOfRecord eventDate institutionCode
2078 384334009 29.51 43.97 HumanObservation 2010-05-20 10:00:00 GeoEcoMar
collectionCode catalogNumber locality
2078 GeoEcoMar BlackSea R/V Mare Nigrum Cruises 2010-2011 GeoEcoMar_BlackSeaCruises_2003_2011_3723 Constanta_10CT05
datasetName phylum order family
2078 Macrobenthos data from the Romanian part of the Black Sea between 2003 and 2011 Mollusca Cardiida Semelidae
genus scientificName originalScientificName scientificNameAuthorship obisID resourceID yearcollected species
2078 Abra Abra alba Abra alba (W. Wood, 1802) 395450 4273 2010 Abra alba
qc aphiaID speciesID continent coordinateUncertaintyInMeters datasetID modified
2078 859307135 141433 395450 Black Sea <NA> IMIS:dasid:5256 2015-12-27 00:00:00
occurrenceID recordedBy scientificNameID class
2078 GeoEcoMar_BlackSeaCruises_2003_2011_3723 <NA> urn:lsid:marinespecies.org:taxname:141433 Bivalvia
lifestage sex individualCount eventID depth minimumDepthInMeters maximumDepthInMeters fieldNumber
2078 <NA> <NA> NA <NA> 60.94 60.94 60.94 I
occurrenceRemarks eventTime footprintWKT identifiedBy
2078 <NA> <NA> <NA> Teaca A.Check points on land
check_onland() uses the xylookup web service which internally uses land polygons from OpenStreetMap to check if any points are located on land. Other shapefiles can be used as well.
check_onland(abra) id decimalLongitude decimalLatitude basisOfRecord eventDate
31 365512845 -0.9092748 54.57467 Occurrence 2011-09-03 10:00:00
institutionCode collectionCode catalogNumber locality
31 Yorkshire Naturalists' Union Marine and Coastal Se 60051 261729389 Skinningrove. Cattersty Sands
datasetName phylum order family genus scientificName
31 Yorkshire Naturalists Union Marine and Coastal Section Records Mollusca Cardiida Semelidae Abra Abra alba
originalScientificName scientificNameAuthorship obisID resourceID yearcollected species qc aphiaID
31 Abra alba (W. Wood, 1802) 395450 3083 2011 Abra alba 1073216639 141433
speciesID continent coordinateUncertaintyInMeters datasetID modified
31 395450 Europe 707.0 IMIS:dasid:3182 2014-04-16 16:16:43
occurrenceID recordedBy
31 urn:catalog:Yorkshire Naturalists' Union Marine and Coastal Se:60051:261729389 Adrian Norris
scientificNameID class lifestage sex individualCount eventID depth
31 urn:lsid:marinespecies.org:taxname:141433 Bivalvia <NA> <NA> NA <NA> NA
minimumDepthInMeters maximumDepthInMeters fieldNumber occurrenceRemarks eventTime footprintWKT identifiedBy
31 NA NA <NA> <NA> <NA> <NA> <NA>check_onland(abra, report = TRUE) field level row message
1 NA warning 31 Coordinates are located on landCheck depth
check_depth uses the xylookup web service to identify which records have potentially invalid depths. Multiple checks are performed in this function:
- missing depth column (warning)
- empty depth column (warning)
- depth values that can't be converted to numbers (error)
- values that are larger than the depth value in the bathymetry layer, after applying the provided depthmargin (error)
- depth values that are negative for off shore points, after applying the provided shoremargin (error)
- minimum depth greater than maximum depth (error)
plot_map(check_depth(abra, depthmargin = 50), zoom = TRUE)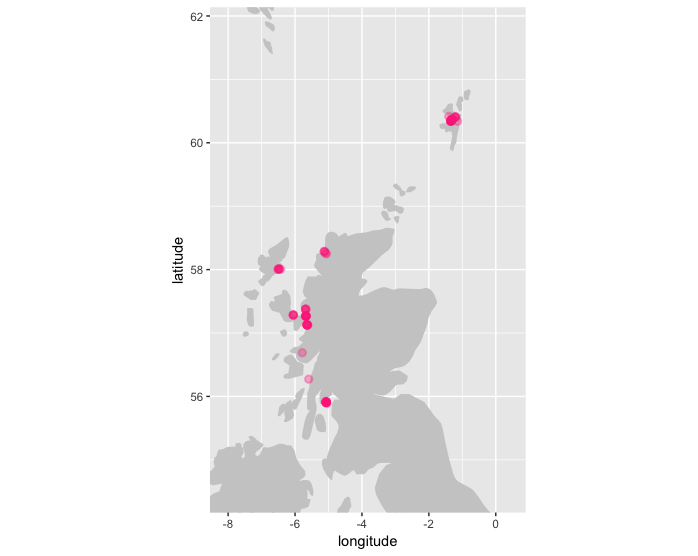
report <- check_depth(abra, report=T, depthmargin = 50)
head(report)field level row message
1 minimumDepthInMeters error 1209 Depth value (52.9) is greater than the value found in the bathymetry raster (depth=-27.0, margin=50)
2 minimumDepthInMeters error 1226 Depth value (62.3) is greater than the value found in the bathymetry raster (depth=4.4, margin=50)
3 minimumDepthInMeters error 1232 Depth value (64.9) is greater than the value found in the bathymetry raster (depth=5.8, margin=50)
4 minimumDepthInMeters error 1235 Depth value (61.2) is greater than the value found in the bathymetry raster (depth=4.0, margin=50)
5 minimumDepthInMeters error 1249 Depth value (68.3) is greater than the value found in the bathymetry raster (depth=8.0, margin=50)
6 minimumDepthInMeters error 1250 Depth value (72.9) is greater than the value found in the bathymetry raster (depth=5.0, margin=50)Check eventID and parentEventID
check_eventids() checks if both eventID() and parentEventID fields are present in an event table, and if all parentEventIDs have a corresponding eventID.
data <- data.frame(
eventID = c("a", "b", "c", "d", "e", "f"),
parentEventID = c("", "", "a", "a", "bb", "b"),
stringsAsFactors = FALSE
)
check_eventids(data) field level row message
1 parentEventID error 5 parentEventID bb has no corresponding eventIDCheck eventID in an extension
check_extension_eventids() checks if all eventIDs in an extension have matching eventIDs in the core table.
event <- data.frame(
eventID = c("cruise_1", "station_1", "station_2", "sample_1", "sample_2", "sample_3", "sample_4", "subsample_1", "subsample_2"),
parentEventID = c(NA, "cruise_1", "cruise_1", "station_1", "station_1", "station_2", "station_2", "sample_3", "sample_3"),
eventDate = c(NA, NA, NA, "2017-01-01", "2017-01-02", "2017-01-03", "2017-01-04", NA, NA),
decimalLongitude = c(NA, 2.9, 4.7, NA, NA, NA, NA, NA, NA),
decimalLatitude = c(NA, 54.1, 55.8, NA, NA, NA, NA, NA, NA),
stringsAsFactors = FALSE
)
occurrence <- data.frame(
eventID = c("sample_1", "sample_1", "sample_2", "sample_28", "sample_3", "sample_4", "subsample_1", "subsample_1"),
scientificName = c("Abra alba", "Lanice conchilega", "Pectinaria koreni", "Nephtys hombergii", "Pectinaria koreni", "Amphiura filiformis", "Desmolaimus zeelandicus", "Aponema torosa"),
stringsAsFactors = FALSE
)
check_extension_eventids(event, occurrence) field level row message
1 eventID error 4 eventID sample_28 has no corresponding eventID in the coreFlatten event records
flatten_event() will recursively add event information from parent events to child events.
event <- data.frame(
eventID = c("cruise_1", "station_1", "station_2", "sample_1", "sample_2", "sample_3", "sample_4", "subsample_1", "subsample_2"),
parentEventID = c(NA, "cruise_1", "cruise_1", "station_1", "station_1", "station_2", "station_2", "sample_3", "sample_3"),
eventDate = c(NA, NA, NA, "2017-01-01", "2017-01-02", "2017-01-03", "2017-01-04", NA, NA),
decimalLongitude = c(NA, 2.9, 4.7, NA, NA, NA, NA, NA, NA),
decimalLatitude = c(NA, 54.1, 55.8, NA, NA, NA, NA, NA, NA),
stringsAsFactors = FALSE
)
flatten_event(event) eventID parentEventID eventDate decimalLongitude decimalLatitude
1 cruise_1 <NA> <NA> NA NA
2 station_1 cruise_1 <NA> 2.9 54.1
3 station_2 cruise_1 <NA> 4.7 55.8
4 sample_1 station_1 2017-01-01 2.9 54.1
5 sample_2 station_1 2017-01-02 2.9 54.1
6 sample_3 station_2 2017-01-03 4.7 55.8
7 sample_4 station_2 2017-01-04 4.7 55.8
8 subsample_1 sample_3 2017-01-03 4.7 55.8
9 subsample_2 sample_3 2017-01-03 4.7 55.8Flatten occurrence and event records
flatten_occurrence() will add event information to occurrence records.
event <- data.frame(
eventID = c("cruise_1", "station_1", "station_2", "sample_1", "sample_2", "sample_3", "sample_4", "subsample_1", "subsample_2"),
parentEventID = c(NA, "cruise_1", "cruise_1", "station_1", "station_1", "station_2", "station_2", "sample_3", "sample_3"),
eventDate = c(NA, NA, NA, "2017-01-01", "2017-01-02", "2017-01-03", "2017-01-04", NA, NA),
decimalLongitude = c(NA, 2.9, 4.7, NA, NA, NA, NA, NA, NA),
decimalLatitude = c(NA, 54.1, 55.8, NA, NA, NA, NA, NA, NA),
stringsAsFactors = FALSE
)
occurrence <- data.frame(
eventID = c("sample_1", "sample_1", "sample_2", "sample_2", "sample_3", "sample_4", "subsample_1", "subsample_1"),
scientificName = c("Abra alba", "Lanice conchilega", "Pectinaria koreni", "Nephtys hombergii", "Pectinaria koreni", "Amphiura filiformis", "Desmolaimus zeelandicus", "Aponema torosa"),
stringsAsFactors = FALSE
)
flatten_occurrence(event, occurrence) eventID scientificName eventDate decimalLatitude decimalLongitude
1 sample_1 Abra alba 2017-01-01 54.1 2.9
2 sample_1 Lanice conchilega 2017-01-01 54.1 2.9
3 sample_2 Pectinaria koreni 2017-01-02 54.1 2.9
4 sample_2 Nephtys hombergii 2017-01-02 54.1 2.9
5 sample_3 Pectinaria koreni 2017-01-03 55.8 4.7
6 sample_4 Amphiura filiformis 2017-01-04 55.8 4.7
7 subsample_1 Desmolaimus zeelandicus 2017-01-03 55.8 4.7
8 subsample_1 Aponema torosa 2017-01-03 55.8 4.7Calculate centroid and radius for WKT geometries
calculate_centroid() calculates a centroid and radius for WKT strings. This is useful for populating decimalLongitude, decimalLatitude and coordinateUncertaintyInMeters.
wkt <- c(
"POLYGON ((2.53784 51.12421, 2.99377 51.32031, 3.34534 51.39578, 2.82349 51.85614, 2.27417 51.69980, 2.53784 51.12421))",
"POLYGON ((3.15582 42.23564, 3.13248 42.14202, 3.22037 42.11249, 3.26019 42.21530, 3.15582 42.23564))"
)
calculate_centroid(wkt) decimalLongitude decimalLatitude coordinateUncertaintyInMeters
1 2.747292 51.50939 45287.041
2 3.193570 42.17716 7531.455Map column names to Darwin Core terms
map_fields() changes column names using a terms map.
data <- data.frame(
id = c("cruise_1", "station_1", "station_2", "sample_1", "sample_2", "sample_3", "sample_4", "subsample_1", "subsample_2"),
date = c(NA, NA, NA, "2017-01-01", "2017-01-02", "2017-01-03", "2017-01-04", NA, NA),
locality = rep("North Sea", 9),
lon = c(NA, 2.9, 4.7, NA, NA, NA, NA, NA, NA),
lat = c(NA, 54.1, 55.8, NA, NA, NA, NA, NA, NA),
stringsAsFactors = FALSE
)
mapping <- list(
decimalLongitude = "lon",
decimalLatitude = "lat",
datasetName = "dataset",
eventID = "id",
eventDate = "date"
)
map_fields(data, mapping) eventID eventDate locality decimalLongitude decimalLatitude
1 cruise_1 <NA> North Sea NA NA
2 station_1 <NA> North Sea 2.9 54.1
3 station_2 <NA> North Sea 4.7 55.8
4 sample_1 2017-01-01 North Sea NA NA
5 sample_2 2017-01-02 North Sea NA NA
6 sample_3 2017-01-03 North Sea NA NA
7 sample_4 2017-01-04 North Sea NA NA
8 subsample_1 <NA> North Sea NA NA
9 subsample_2 <NA> North Sea NA NACheck eventDate
data <- data.frame(eventDate = c(
"2016",
"2016-01",
"2016-01-02",
"2016-01-02 12",
"2016-01-02 12:34",
"2016-01-02 12:34:48",
"2016-01-02T13:00:00+01:00",
"2016-01-02T13:00:00+0100",
"2016-01-02T13:00:00+01",
"2016-01-02T13:00:00Z",
"2016-01-02 13:00:00/2016-01-02 14:00:00",
"2016-01-02 13:00:00/14:00:00",
"2016-01-02/05",
"2016-01-01 13u40",
"2016/01/03",
"",
NA
), stringsAsFactors = FALSE)
check_eventdate(data) level row field message
1 error 14 eventDate eventDate 2016-01-01 13u40 does not seem to be a valid date
2 error 15 eventDate eventDate 2016/01/03 does not seem to be a valid date
3 error 16 eventDate eventDate does not seem to be a valid date
4 error 17 eventDate eventDate NA does not seem to be a valid dateData quality report
report() generates a basic data quality report.
report(abra)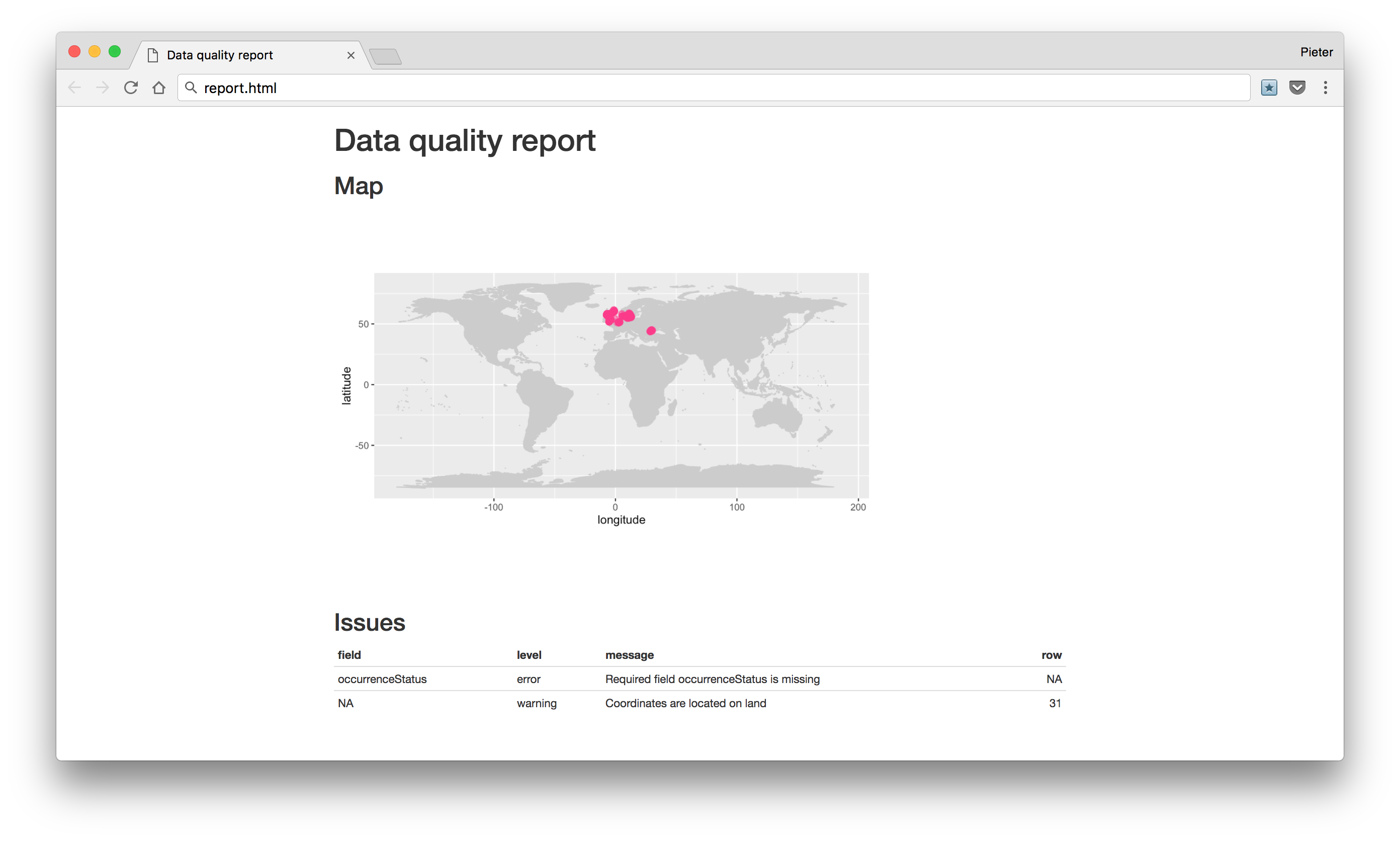
Lookup XY
lookup_xy returns basic spatial information for a given set of points. Currently three main things are returned: information from vector data (areas), information from rasters (grids) and the shoredistance.
xydata <- lookup_xy(abra, shoredistance = TRUE, grids = TRUE, areas = TRUE)
head(xydata)shoredistance sstemperature sssalinity bathymetry obis
1 30 10.28631 34.76271 -4.0 United Kingdom, United Kingdom, F, T, eez, eez, United Kingdom: all, United Kingdom, United Kingdom: all, 221
2 1080 10.33242 34.90622 61.4 United Kingdom, United Kingdom, T, F, eez, eez, United Kingdom, United Kingdom: all, 221, United Kingdom: all
3 1184 10.72199 34.88896 122.2 United Kingdom, United Kingdom, T, F, eez, eez, United Kingdom, United Kingdom: all, 221, United Kingdom: all
4 290 10.79197 34.29342 20.6 United Kingdom, United Kingdom, F, T, eez, eez, United Kingdom: all, United Kingdom, United Kingdom: all, 221
5 259 10.72199 34.88896 51.0 United Kingdom, United Kingdom, F, T, eez, eez, United Kingdom: all, United Kingdom, United Kingdom: all, 221
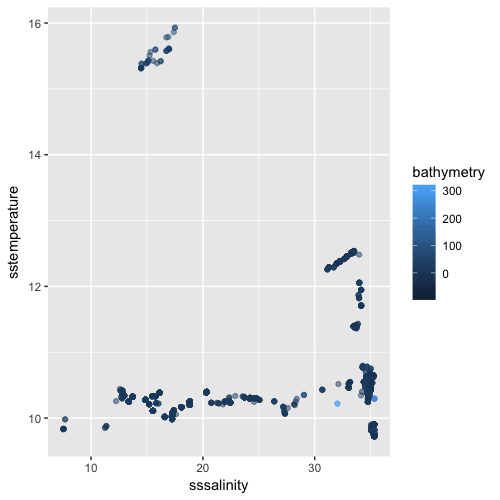
6 506 10.77101 34.30700 32.4 United Kingdom, United Kingdom, F, T, eez, eez, United Kingdom: all, United Kingdom, United Kingdom: all, 221ggplot(xydata, aes(sssalinity, sstemperature, color=bathymetry)) +
geom_point(alpha=0.5)