Can you look at the kernel.json in each of those kernelspec directories you found?
Open rjtjs opened 7 years ago
Can you look at the kernel.json in each of those kernelspec directories you found?
kernel.json in python2 directory:
{
"display_name": "Python 2",
"language": "python",
"argv": [
"python",
"-m",
"ipykernel_launcher",
"-f",
"{connection_file}"
]
}kernel.json in python3 directory:
{
"argv": [
"python",
"-m",
"ipykernel_launcher",
"-f",
"{connection_file}"
],
"display_name": "Python 3",
"language": "python"
}This looks similar to jupyter/jupyter#270 . Ping @minrk - I think this is due to installing kernelspecs automatically as data files in the wheel.
For now, you can probably work around it by changing the first item in argv to 'python3' in the Python 3 kernel.json file.
Yup, it's the same. Running ipython kernel install for Python 2 and 3 will resolve the ambiguity by installing with sys.executable's abspath that we can't do in data_files. While the new data_files work better in envs of all kinds and with just one Python, two Pythons running on the same prefix (e.g. --user on linux, but not mac) run into ambiguity.
I didn't fully understand what @minrk said above, but running ipython kernel install --name python3 didn't solve the issue, if that helps.
As @takluyver suggested, changing argv solved it for me, thank you. I changed the first argument of argv in python3 folder's kernel.json from python2 path to python3 path, and everything works as expected.
I got a similar problem
I solved it by sudo pip install --upgrade ipykernel
Solved this issue by executing
python3 -m ipykernel install --userI also am experiencing this issue, and editing the two kernel.json files has solved the problem. Will I have to do this every time I upgrade jupyter on my machine?
Hopefully not. We've refined this behaviour in a new version of jupyter_client
This is also happening to me. Using ipython3 notebook works.
Solved this issue by executing
python3 -m ipykernel install --user
works like charm
My jupyter notebook was using a python3 kernel even though I wanted it to use a python2 kernel (and the web interface was showing python2).
import platform
print(platform.python_version())jupyter kernelspec list
sudo -H pip2 install jupyter
This was my solution: https://github.com/jupyter/notebook/issues/2359#issuecomment-453205175
works for me with python3 -m ipykernel install --user - THANK YOU nvdv 👍
Changing the executable path of jupyter (in the kernel.json) worked in my case.
For details see https://stackoverflow.com/a/57274684/3521099 .
I'm having a similar issue on Mac OS. When trying to run the kernel within a virtual environment, the system python (brew installed) is loaded instead.
When the virtualenv is active:
(venv) lubaroli@machine: ~$ which python
/Users/lubaroli/.env/venv/bin/pythonLikewise:
(venv) lubaroli@machine: ~$ which IPython
/Users/lubaroli/.env/venv/bin/IPythonWhen running:
python kernel install --user --name=venv
The kernel.json file reads:
{
"argv": [
"/usr/local/opt/python/bin/python3.7",
"-m",
"ipykernel_launcher",
"-f",
"{connection_file}"
],
"display_name": "venv",
"language": "python"
}Changing the kernel.json path manually to the venv path does not fix the issue. When running a notebook, sys.executable still returns:
'/usr/local/opt/python/bin/python3.7'
Any ideas?
Solved mine with python3 -m ipykernel install --user
as reported by others, sys.path and sys.executable agree from within the virtualenv (I'm using pipenv, btw), the latter correctly pointing to the virtualenv:
>>> sys.executable
'/Users/tullio/.local/share/virtualenvs/data_science-V36xIoci/bin/python'but from within jupyter lab, sys.executable moves to pointing to
>>> sys.executable
'/usr/local/opt/python/bin/python3.7'so far, 2 solutions have been proposed, neither of which worked for me:
python3 -m ipykernel install --userdid not fix the issue for me (I'm assuming it has to be run after activating the venv, and before opening jupyter lab, right?)
kernel.json fileRunning
jupyter kernelspec listreturns me the same, single kernel.json file regardless of whether I do it from the virtualenv or from the jupyter lab instance I launch from within it. argv[0] really looks fine to me, no reason to change it:
$ more kernel.json
{
"argv": [
"/Users/tullio/.local/share/virtualenvs/data_science-V36xIoci/bin/python3",
"-m",
"ipykernel_launcher",
"-f",
"{connection_file}"
],
"display_name": "Python 3",
"language": "python"
}And seriously, jupyter: threads like this one have been going on for ages. I've installed jupyter lab from within the virtualenv - WHY ON EARTH would I want sys.executable to point to anywhere but to the same value that will be returned when launching python from the virtualenv?
I opened /usr/local/share/jupyter/kernels/python2/kernel.json and rewrite the python path, but I couldn't save the file after that.
I am using a server instead of stand alone OS, just in case this is the reason, if it is like this, do you have any idea please?
The inability to save edits to a file directly is more of a system thing. You'll need to look at the kernel.json's permissions and change users and/or permissions to complete the save operation. Since this is a server, you may need to enlist the help of your system's administrator.
Instead of changing kernel under /usr/local/share/jupyter/kernels/python3/kernel.json, the problem solved by modifying the kernel under ~/.local/share/jupyter/kernels/python3/kernel.json into { "argv": [ "home/username/anaconda3/bin/python3", //username here refers to specific's user name "-m", "ipykernel_launcher", "-f", "{connection_file}" ], "display_name": "Python 3", "language" : "python" } Just if it could help anyone else who encounter the same problem as I do.
solved my problem with python3 -m ipykernel install --user on macOS Catalina 10.15.3 with anaconda
same issue here as tbagnoli and lubaroli using conda environments.
I generate the environment using the code below:
conda create --name venv python pip ipykernel
conda activate venv
pip install -e /path/to/local/packageWithin terminal the environments agree:
~
venv ❯ which python
/Users/nicholasmi/opt/anaconda3/envs/venv/bin/python
~
venv ❯ which pip
/Users/nicholasmi/opt/anaconda3/envs/venv/bin/pipI then install the kernelspec via:
~
venv ❯ python -m ipykernel install --user --name venv --display-name "Python (venv)"Using the kernelspec in Jupyter then yields:

Does a solution exist? The methods above don't help. For example, the kernel.json file seems to be set up correctly already.
~/Library/Jupyter/kernels/venv 19s
base ❯ cat kernel.json
{
"argv": [
"/Users/nicholasmi/opt/anaconda3/envs/venv/bin/python",
"-m",
"ipykernel_launcher",
"-f",
"{connection_file}"
],
"display_name": "Python (venv)",
"language": "python"
}```I just recently started to get the exact same behaviour as @nicholas-miles - glad it's not only me.
Were you in the activated environment (e.g., venv) when you launched the notebook server?
I use conda for my venvs, but yes.
And doing which jupyter confirmed that I was calling the jupyter executable that's in my conda venv path.
Thanks @mathieuboudreau. I had seen that the kernel.json file was viewed from the base env in @nicholas-miles' example, so I thought I'd toss that out there.
When you say you recently started to get the exact same behavior, how recent are you talking about and were there any significant upgrades you may be aware of around that time as well?
I was on vacation last week. If I recall, it was working correctly before then, and isn't working now. I don't think made any upgrades, but I could try and delete anaconda and reinstall it.
I got the same issue!
For some reason the environment is not activated in jupyter notebook. If I activate the environment in the terminal conda activate myenv and run echo $PATH, I get /Users/$USER/anaconda3/envs/myenv/bin.
However, when I start the jupyter notebook and run !echo $PATH, I get /Users/$USER/anaconda3/bin.
Therefore, also if I run which pythonin the terminal I get the python version from the environment but if I run it in the notebook I get the default python.
This is on Mac Catalina.
You might find something helpful here and in the referenced links.
Tested everything on https://ipython.readthedocs.io/en/stable/install/kernel_install.html#kernels-for-different-environments still having the same issue as @nicholas-miles
for the love of GOD, somebody please address this issue, none of the solutions work, and many have the same issue where changing the kernel does not point the python to the correct kernel environment. For me it always point into the same environment from which i ran the initial jupyter-notebook. I am stuck to that environment python, cannot change the kernel to any other one. Within jupyter if i run this:
import sys
sys.executableit points to the correct python environment and changes as expected, but
which python
inside the same damn jupyter notebook always point to the one directory even when i change the kernel.
It seems like the crux of the issue is that the correct python is in use by the kernel, but the PATH (from the kernel) doesn't reflect the python in use. As a result, while python statements (e.g., import foo) produce the desire results, any shell commands (e.g., !pip install foo) do not. In such cases, you could fully-qualify the shell command and, I believe, get the desire results.
I found the IPython's documentation link in the Python Environments section of the troubleshooting documentation to be fairly helpful. In particular the portion about installing the kernel with the --prefix option to point at an env different that the current env.
Jupyter, by default, doesn't activate environments for a given kernel. This is why the PATH-based commands (sans prefixing) can be problematic. If you find that activation may resolve things for you, then perhaps taking a look at nb_conda_kernels may be helpful. This package updates your jupyter's configuration to use an alternate KernelSpecManager that is used for discovering kernels (which some find invasive). These kernel specifications contain information that, AFAIK, activate the environment associated with that kernel prior to its launch.
Here's a screenshot of a kernel I've started that uses a python from an environment other than the notebook server. The notebook server is launched in my activated notebook-dev env, while the kernel I want to run is in my enterprise-gateway-dev env. Aside from the PATH and sys.executable difference, you'll find that the import statement produces the desired result (my notebook-dev env doesn't have kubernetes installed). In addition, my EG env doesn't have nose installed until I fully qualify the pip command to install it into the kernel's env (i.e., enterprise-gateway-dev).

I hope you find this helpful and apologize for the frustration.
It seems like the crux of the issue is that the correct python is in use by the kernel, but the PATH (from the kernel) doesn't reflect the python in use. As a result, while python statements (e.g.,
import foo) produce the desire results, any shell commands (e.g.,!pip install foo) do not. In such cases, you could fully-qualify the shell command and, I believe, get the desire results.
I found this exact behavior when trying having jupyterlab to manage multiple env.
kernel.json, each pointing different env.import behave exacty as expected, only import from the pointed env!system command including !pip install and python3 --version will only point to the main kernel (in my case, default ubuntu python 3.8)Is there any work around to install package diractly to env without leaving jupyterlab?
Hi @yasirroni - try using the %pip magic. This is exactly the problem it resolves.
ipython kernel install --user worked for me

What do I need to do to install dependency?
run pip install pandas, it installed to /usr/bin/python2 instead of /opt/python3.9
@jianboy I solved it with this
run pip install pandas, it installed to /usr/bin/python2 instead of /opt/python3.9
import sys
! {sys.executable} -m pip install pandas
import pandasYou may need to restart the kernel
The problem with which python inside notebook not pointing to the same as sys.executable is caused by a difference in system paths. In order to make sure they are pointing to the same executable:
python -m ...--env PATH=...For my setup with virtualenv it looks like this
/home/user/venv/bin/python -m pip install ipython ipykernel
/home/user/venv/bin/python -m ipykernel install --user --name=kernel-name --env PATH /home/user/venv/bin:$PATHIn general:
<YOUR_PYTHON_EXECUTABLE> -m pip install ipython ipykernel
<YOUR_PYTHON_EXECUTABLE> -m ipykernel install --user --name=kernel-name --env PATH <YOUR_PYTHON_BIN_DIRECTORY>:$PATH@nemusa Thank you so much, this worked for me! It was driving me crazy that I couldn't figure out why the PATH inside the shell didn't match my $PATH
@nemusa 's approach was the only thing that worked for me running the jupyter/minimal-notebook docker contaner
I am on OSX Sierra (10.12.5), and am using Jupyter Notebook v 4.3.0. I have installed both Python2 and Python3 using Homebrew, and both kernels for Jupyter Notebook. Upon opening the notebook to use Python3 kernel, I find that it still uses Python2.
I might be missing something obvious in my installation, but I can't seem to figure out what it is.
Detailed Description
Installation:
I installed both Python2 and Python3 using Homebrew, and Jupyter Notebook on my system thus:
brew install python3pip3 install --upgrade pip3 setuptools wheelpip3 install jupyterI ran all analogous commands for Python2 as well.
Configuration:
I generated the config file using
jupyter notebook --generate-config, and the only configurations I have "enabled" in my config file are:c.NotebookApp.browser = u'safari'c.NotebookApp.password = 'sha1:somepwd'Both kernels installed:
Both kernels seem to be visible to kernelspec:
Both kernels are also available as options when I start execute
jupyter notebook:As an additional check,
pip listandpip3 listboth includeipykernel.Problem:
When I opened a new notebook using Python3 kernel, none of the packages I installed using
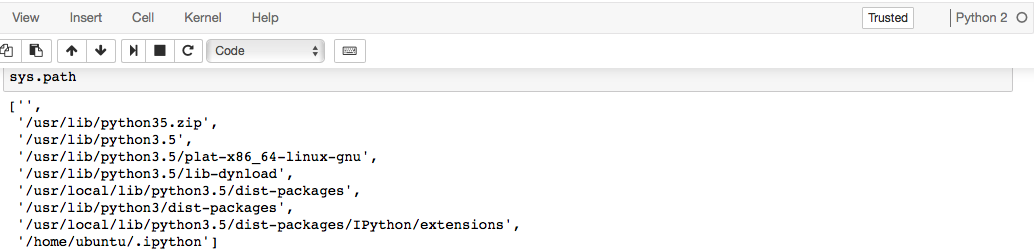
pip3were recognized when I usedimport. Furthermore,sys.pathreturns path to Python2:When I run
print "Hello World", it gives me a successful output (which is true in Python2, but should throw an error in Python3).However, when I use the (depreciated) command
ipython3 notebook, things work as expected:and all my Python3 packages are successfully recognized.
It doesn't seem to be the biggest deal breaker, as I can continue working with
ipython3 notebook, but I wonder what I am missing that causesjupyter notebookto not load Python3.