Hi Eleonora,
Provided:
- the data are registered --- i.e., spatially aligned, and
- you use nonparametric inference
then yes, using spm1d is valid.
Considering these two issues separately...
- Registration
The data must be spatially aligned. Consider this figure which depicts means for two conditions:
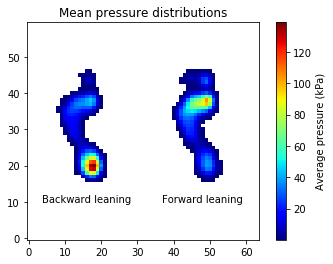
Notice that the means retain the original foot shapes. If the data were not spatially registered, mean images like that would contain strange looking foot distortions.
2D data registration is not supported in spm1d, so you'd need to register your data elsewhere before using spm1d.
- Nonparametric inference
Parametric inference in spm1d is limited to 1D data; these parametric procedures estimate both 1D smoothness and 1D topological features like the Euler characteristic --- which is basically the number of supra-threshold clusters. For parametric analysis of 2D data one would need to implement 2D smoothness estimates and 2D topological feature estimates. These do not exist in spm1d but do exist in other packages including, for example: SPM12
Fortunately, calculating the critical threshold nonparametrically does not require these 2D calculations. For nonparametric inference, the critical threshold is based on the maximum test statistic value across the entire domain, regardless of whether the domain is 1D, 2D or nD. Note that flattening nD data into a 1D vector format does not affecting the maximum statistic value. Nonparametric inference in spm1d works by randomly permuting the observations, and building a numerical distribution of the maximum test statistic value, and it is this distribution from which the critical threshold is calculated, usually as the 95th percentile.
However, spm1d's cluster-level probabilities are NOT valid for 2D data. Calculating cluster-level p values for 2D data requires 2D geometry calculations (e.g. cluster area), and these are not available in spm1d.
A complete working example for 2D analysis is available here: https://spm1d.org/doc/Stats2D/ex2d.html Note that these data are pre-registered, and that only nonparametric inference is used.
Hi Todd,
I was wondering if you could clarify to me the use of SPM1D to analyse 2D pressure data, in particular the conditions that allow to undertake the SPM1D analysis onto 2D data sets.
Thanks, Eleonora