Your interpretations are correct except for: "...it will perform a t-test at each point". Let's first consider the case of a single test (e.g. a two-sample t test) before considering post hoc tests.
For a single test it is more accurate to think of SPM's calculations like this:
- SPM calculates the test statistic value (t-value, F-value, etc.) at each point, then
- SPM conducts one test at the whole-continuum level.
Just as calculating a mean and a standard deviation (SD) does not represent a "test", calculating a test statistic value (which is basically just the ratio between the mean effect and the SD) also does not represent a test. A test is conducted only when probability calculations are made.
Another relevant interpretation is based on the figure below (from this site). In this figure the "FWHM" parameter represents smoothness, where FWHM = 0 represents perfectly rough, uncorrelated data and where FWHM = $\infty$ represents perfectly smooth data (i.e., flat lines). For purposes of discussion let us say the there are Q points that have been used to characterize the continuum, where Q is often 100 or 101 in real data analysis.
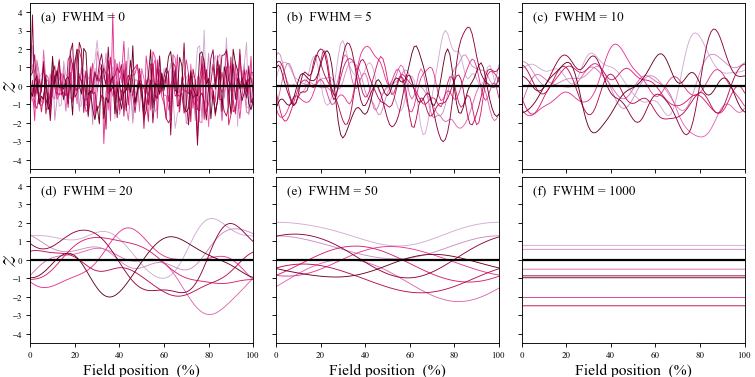
Your interpretation is correct for the case of FWHM = 0, when each of the Q points along the continuum effectively embodies an independent random process. In this case you correctly point out that the Bonferroni correction can be used to accurately control Type I error across the Q independent tests.
At the other extreme, when the data are perfectly smooth ( FWHM = $\infty$ ) there is effectively just a single random process, as all Q points are perfectly correlated with each other. In this case the usual procedures found in many standard software packages like SPSS and MATLAB can be used to accurately control Type I error.
SPM and SPM-related corrections deal with the intermediary case of 0 < FWHM < $\infty$. In this case there is effectively more than one random process but fewer than Q random processes. SPM uses smoothness (in the form of gradient estimates) to estimate the effective number of random processes and thereby control Type I error. Note that using no correction would yield greatly increased Type I error, and using a Bonferroni correction would yield a greatly increased Type II error. Only SPM and SPM-like methods (like those in the Functional Data Analysis literature) can be used to accurately control both Type I and Type II error for case of 0 < FWHM < $\infty$.
A key point is that SPM results are invariant to Q. Sampling the data at Q=100, 1000, or 1 million points greatly affects Bonferroni-corrected results but does not affect SPM results.
Please see this link at spm1d.org for a related consideration.
Finally, to return to your question about post hoc tests...
For post hoc tests there are two levels of corrections:
- Continuum-level corrections
- Multiple comparisons corrections
Continuum-level corrections are those described above. Multiple comparisons corrections are used whenever more than one test is conducted, for example in ANOVA post hoc testing, but also for the case of multiple dependent variables like four separate joint angles. Bonferroni and related corrections can indeed be used for multiple comparisons corrections like these. SPM or related methods should be used for continuum-level corrections. As far as I am aware there is no established or standard methodology for post hoc tests in the SPM and related literatures, and that typical corrections like the Bonferroni correction can be used.





Hello everyone,
I want to compare a markerless motion capture system to a marker based system and to a IMU based system. For that purpose, I investigate the kinematics during Gait and Squat, probably evaluating a total of 4 joint angles. The data have been captured simultanouesly. I want to normalize the data of each subject to a graph between 0-100% and then compare these values. My approach to identify differences between these systems would be to run multiple SPM ANOVA's for each angle with subsequent post-hoc paired t-tests.
For that analysis I have: 3 factors: Markerless, Marker-Based, IMU 4 dependent variables: 4 Angles
My question: If I understand the post-hoc SPM approach correctly, it will perform a t-test at each point of the normalized graph, resulting in a huge amount of t-tests. Won't this inevitably lead to a type 1 error? As far as I know, the Bonferroni adjustment can be used to help with this problem, which in my case would be alpha = 0.05/3, = 0.0166. Still, is this correction sufficient or are there other ways which would be more appropriate to counteract the type 1 error?
I'd greatly aprecciate your help and thoughts on my approach. Thank you!