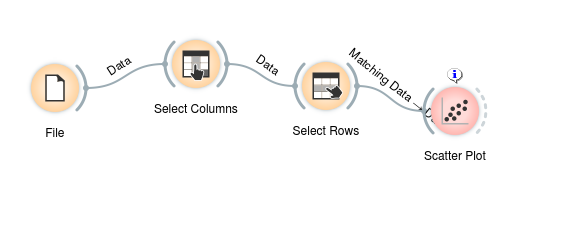
I started off by trying to find a way to manually insert inputs into the widgets that were generated during code generation. As it turns out, that isn't necessary at all and by removing some hacky code that I inserted as I was still learning the Orange codebase, I was able to give access to the actual widget instances from the scheme rather than having to create new, blank ones.
Using this newly acquired capability, I wrote code generators for owselectrows and owselectcolumns as well as creating some __repr__ functions for two Orange.data.filter classes.
┆Issue is synchronized with this Asana task