The whole picture won't look good for some obvious reasons, but, after zooming in, the results look similar to the ones we're producing (albeit with much more effort and less scalability):
After zooming:
Let's try to improve these results and replace our current time-series histogram approach with this one.
The following seems to demonstrate how we can use
datashaderto produce distribution plots rather quickly for large series:The whole picture won't look good for some obvious reasons, but, after zooming in, the results look similar to the ones we're producing (albeit with much more effort and less scalability):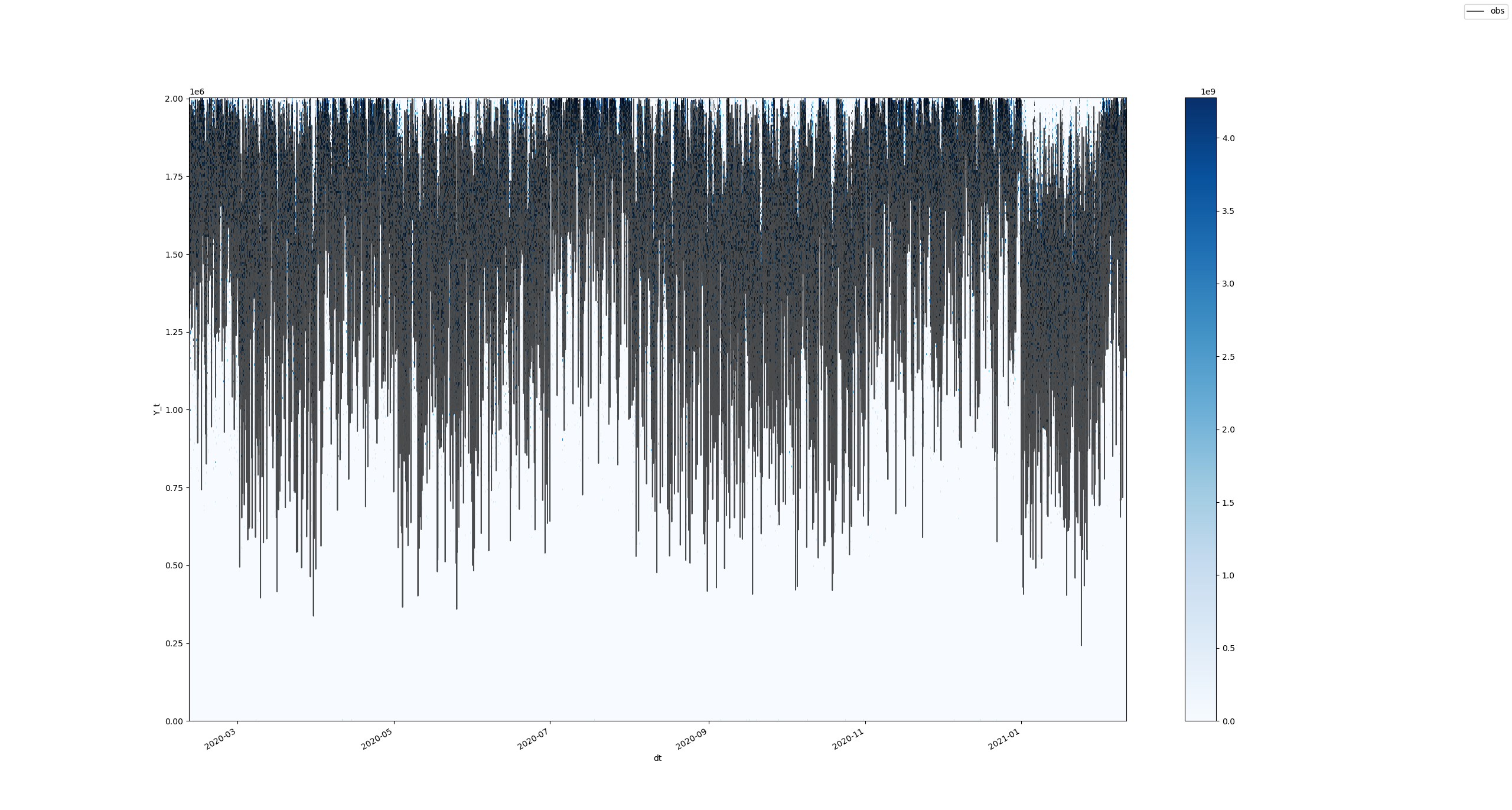
After zooming: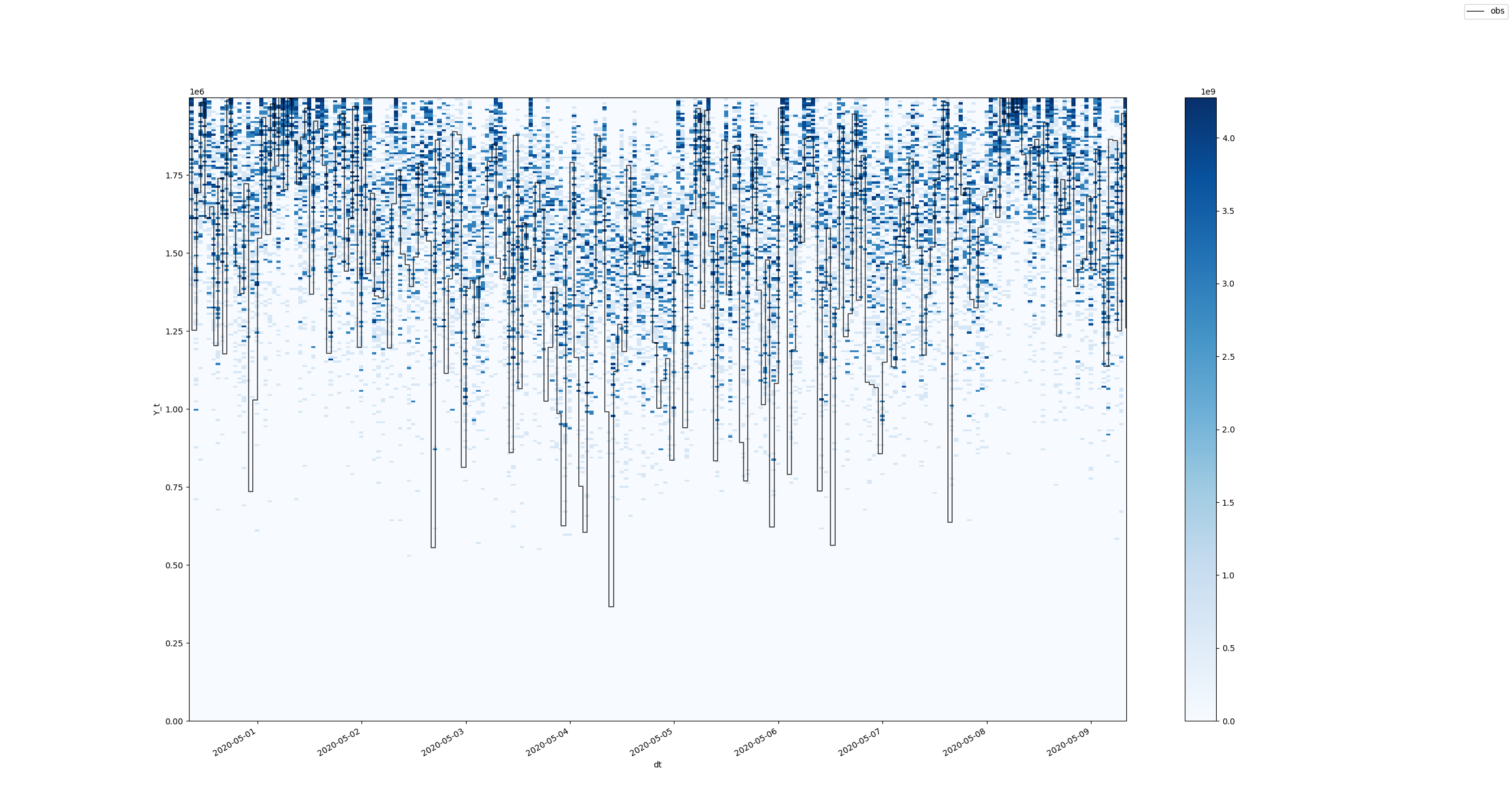
Let's try to improve these results and replace our current time-series histogram approach with this one.