Closed avirshup closed 7 years ago
Regression after merging the no-RPC stuff. After making a selection:
`rs = mdt.widgets.ResidueSelector(mol) rs``
Expected: Before the merge, rs.selected_atoms would yield a list of atoms, and rs.selected_residues would yield a list of residues:
rs.selected_atoms
rs.selected_residues
>>> rs.selected_atoms [<Atom HO5' (elem H), index 0 (res DA50 chain 1) ... >, <Atom O5' (elem O), index 1 (res DA50 chain 1) in ...>, ... ] >>> rs.selected_residues [<Residue DA50 (index 0, chain 1) in Molecule: Unnamed molecule from OpenMM>]
Getting: rs.selected_residues returning a list of strings; rs.selected_atoms doesn't appear to be working.
>>> rs.selected_atoms AttributeError: selected_atoms >>> rs.selected_residues ['DA50']
Fixed over here: https://github.com/Autodesk/molecular-design-toolkit/pull/106
Regression after merging the no-RPC stuff. After making a selection:
`rs = mdt.widgets.ResidueSelector(mol) rs``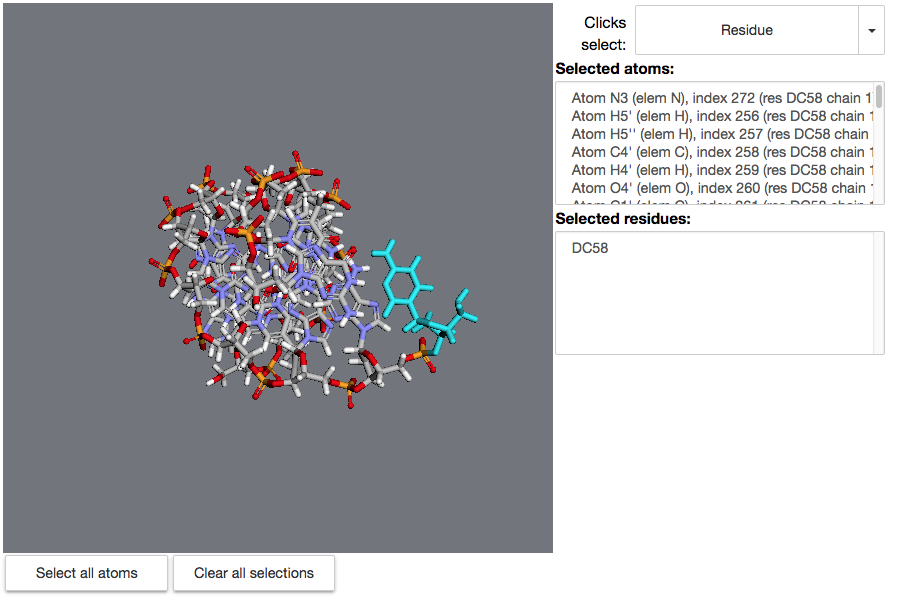
Expected: Before the merge,
rs.selected_atomswould yield a list of atoms, andrs.selected_residueswould yield a list of residues:Getting:
rs.selected_residuesreturning a list of strings; rs.selected_atoms doesn't appear to be working.