Thank you @choc2000 for the input.
I have replaced the line
tmpFacet = as.numeric(gsub("\\D", "", tmpStrata))
with
tmpFacet = gsub(".*=", "", tmpStrata)
which keep the level values for each strata and facet factor as it is.
If we would have tmpStrata as "sex=A","ph.ecog=0",
the previous code just screening for numbers and it would produce tmpFacet as N/A, 0, and
the new code would produce A, 0.
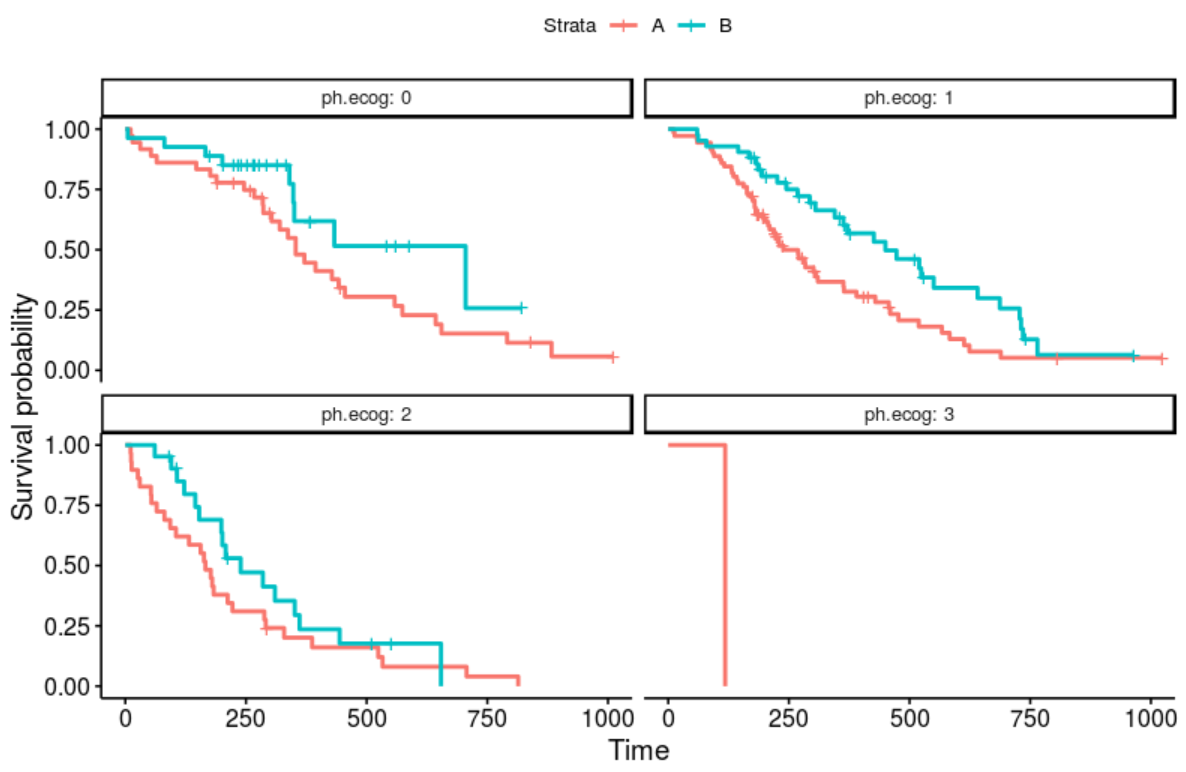
Your test would produce a plot as follows:
lung$sex <- LETTERS[lung$sex]
fit <- survfit(Surv(time, status) ~ sex, lung)
ggsurvplot_facet_fix(fit, lung, facet.by="ph.ecog")
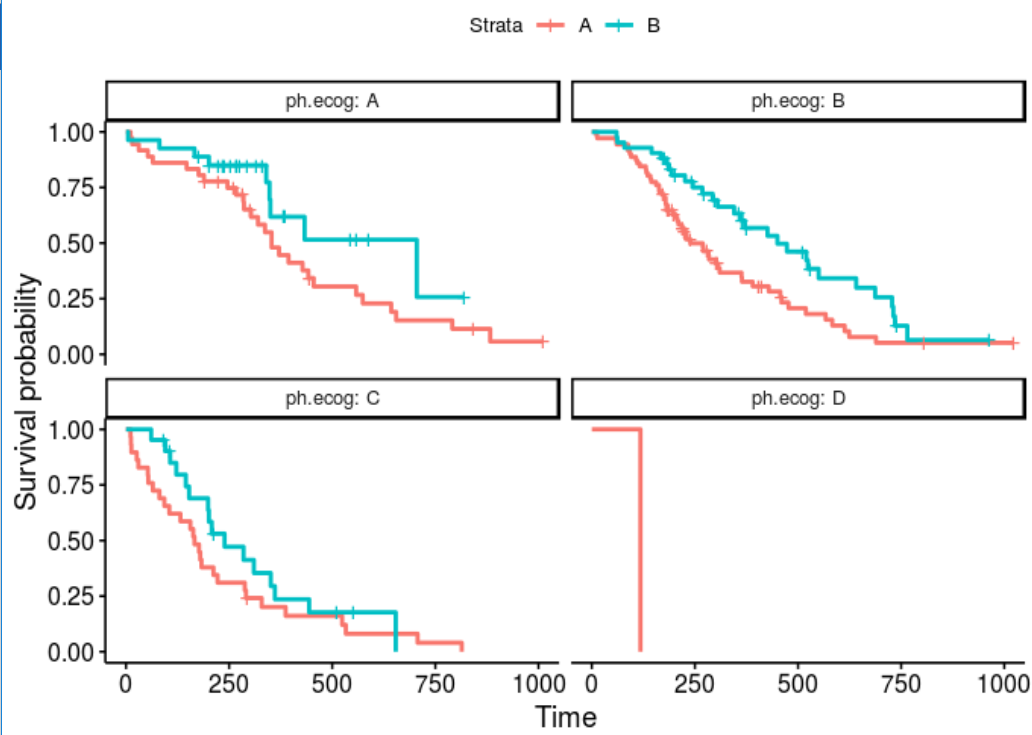
We can also try to make strata and facet factor is characters:
lung$sex <- LETTERS[lung$sex]
lung$ph.ecog <- LETTERS[lung$ph.ecog+1]
fit <- survfit(Surv(time, status) ~ sex, lung)
ggsurvplot_facet_fix(fit, lung, facet.by="ph.ecog")
Seems like this fix only works if the selected strata are numeric in the original data. I guess, this is due to the following line of code in the
.connect2origin_fixfunction:tmpFacet = as.numeric(gsub("\\D", "", tmpStrata))This line replaces all non-numeric characters from the strata.Try the following in your example with the
lungdata set before fitting the model:lung$sex <- LETTERS[lung$sex]In this case, the line above yieldsNAas strata, and the resulting plot looks like this:Please note that survival curves are still not connected to the origin, but instead a third strata
NAis introduced (which does not show up in the plots as there are no data points belonging to this group except for the newly created ones in the origin).