2. CUDA implementation
Storategy
- Generate poisson and normal random numbers in a kernel.
- Use of
curand_poissonandcurand_normalofcurand.
- Use of
Verification
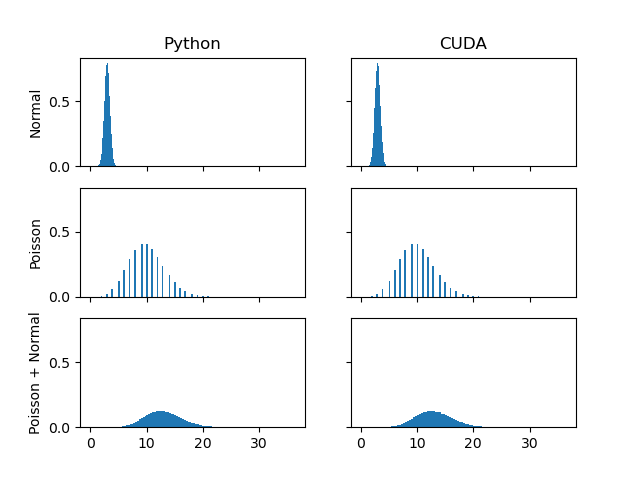
- Check the distribution of the gerated numbers are the same between CUDA and python.
Code
test_curand.pyimport time import numpy as np import matplotlib.pyplot as plt import tigre import tigre.algorithms as algs from tigre.utilities import sample_loader import tigre.utilities.gpu as gpu
import _RandomNumberGenerator as RNG
This is just a basic example of very few TIGRE functionallity.
We hihgly recomend checking the Demos folder, where most if not all features of tigre are demoed.
listGpuNames = gpu.getGpuNames() if len(listGpuNames) == 0: print("Error: No gpu found") else: for id in range(len(listGpuNames)): print("{}: {}".format(id, listGpuNames[id]))
gpuids = gpu.getGpuIds(listGpuNames[0]) print(gpuids)
Geometry
geo = tigre.geometry(mode="cone", nVoxel=np.array([256, 256, 256]), default=True) geo.dDetector = np.array([0.8, 0.8]) 2 # size of each pixel (mm) geo.sDetector = geo.dDetector geo.nDetector
print(geo)
nangles = 512 angles = np.linspace(0, 2 * np.pi, nangles, endpoint=False, dtype=np.float32)
fig, axes = plt.subplots(3,2, sharex=True, sharey=True)
list_results = []
####################
GAUSSIAN NOISE
#################### projections_g = np.zeros([nangles, geo.nDetector[0], geo.nDetector[1]], dtype=np.float32) Gaussian = np.array([3, 0.5])
noise_data = np.random.normal(Gaussian[0], Gaussian[1], size=projections_g.shape) # warmup
time_start = time.time() noise_data = np.random.normal(Gaussian[0], Gaussian[1], size=projections_g.shape) time_end = time.time()-time_start print(noise_data.shape) list_results.append("G0 time: {}, mean: {}, sd : {}, min: {}, max: {}".format(time_end, np.average(noise_data), np.sqrt(np.var(noise_data)), np.min(noise_data), np.max(noise_data))) noise_data = noise_data.reshape([-1]) ax = axes[0,0] ax.set_title("Python") ax.set_ylabel("Normal") count, bins, ignored = ax.hist(noise_data, 100, density=True)
time_start = time.time() noise_data = RNG.add_noise(projections_g, Gaussian[0], Gaussian[1]) time_end = time.time()-time_start list_results.append("G1 time: {}, mean: {}, sd : {}, min: {}, max: {}".format(time_end, np.average(noise_data), np.sqrt(np.var(noise_data)), np.min(noise_data), np.max(noise_data))) noise_data = noise_data.reshape([-1]) ax = axes[0,1] ax.set_title("CUDA") count, bins, ignored = ax.hist(noise_data, 100, density=True)
####################
POISSON NOISE
####################
projections_p = np.zeros([nangles, geo.nDetector[0], geo.nDetector[1]], dtype=np.float32) projections_p[:, :, :] = 10 noise_data = np.random.poisson(projections_p) #warmup
time_start = time.time() noise_data = np.random.poisson(projections_p) time_end = time.time()-time_start list_results.append("P0 time: {}, mean: {}, sd : {}, min: {}, max: {}".format(time_end, np.average(noise_data), np.sqrt(np.var(noise_data)), np.min(noise_data), np.max(noise_data))) noise_data = noise_data.reshape([-1]) ax = axes[1, 0] ax.set_ylabel("Poisson") count, bins, ignored = ax.hist(noise_data, 100, density=True)
rng = np.random.default_rng()
time_start = time.time()
noise_data = rng.poisson(projections_p)
time_p[1] = time.time()-time_start
print("P2 mean: {}, sd : {}".format(np.average(noise_data), np.sqrt(np.var(noise_data))))
noise_data = noise_data.reshape([-1])
count, bins, ignored = axes[1, 1].hist(noise_data, 100, density=True)
noise_data = RNG.add_poisson(projections_p) #warmup
time_start = time.time() noise_data = RNG.add_poisson(projections_p) time_end = time.time()-time_start list_results.append("P1 time: {}, mean: {}, sd : {}, min: {}, max: {}".format(time_end, np.average(noise_data), np.sqrt(np.var(noise_data)), np.min(noise_data), np.max(noise_data))) noise_data = noise_data.reshape([-1]) ax=axes[1, 1] count, bins, ignored = ax.hist(noise_data, 100, density=True)
####################
POISSON +GAUSSIAN
####################
noise_data = np.random.poisson(projections_p) # warmup
time_start = time.time() noise_data = np.random.poisson(projections_p) noise_data = noise_data + np.random.normal(Gaussian[0], Gaussian[1], size=noise_data.shape) time_end = time.time()-time_start list_results.append("PG0 time: {}, mean: {}, sd : {}, min: {}, max: {}".format(time_end, np.average(noise_data), np.sqrt(np.var(noise_data)), np.min(noise_data), np.max(noise_data))) noise_data = noise_data.reshape([-1]) ax = axes[2, 0] ax.set_ylabel("Poisson + Normal") count, bins, ignored = ax.hist(noise_data, 100, density=True)
noise_data = RNG.add_noise(projections_p, Gaussian[0], Gaussian[1]) # warmup
time_start = time.time() noise_data = RNG.add_noise(projections_p, Gaussian[0], Gaussian[1]) time_end = time.time()-time_start list_results.append("PG1 time: {}, mean: {}, sd : {}, min: {}, max: {}".format(time_end, np.average(noise_data), np.sqrt(np.var(noise_data)), np.min(noise_data), np.max(noise_data))) noise_data = noise_data.reshape([-1]) ax=axes[2, 1] count, bins, ignored = ax.hist(noise_data, 100, density=True)
####################
Show Results
#################### for line in list_results: print(line)
plt.savefig("CompareCudaAndPython.png") plt.show()
exit()
### Result
* Distributions look the same
* CUDA version is faster in most of the cases.
G0 time: 0.796917200088501, mean: 3.0000234450711574, sd : 0.4999682128226512, min: 0.3364493374464943, max: 5.64485169303217 G1 time: 0.9529380798339844, mean: 3.0000410079956055, sd : 0.49989402294158936, min: 0.21702957153320312, max: 5.765656471252441 P0 time: 2.7501330375671387, mean: 9.99996104836464, sd : 3.1621095114201148, min: 0, max: 32 P1 time: 0.07698988914489746, mean: 10.00020980834961, sd : 3.1637797355651855, min: 0.0, max: 31.0 PG0 time: 3.5634419918060303, mean: 13.000713100842493, sd : 3.2016939344581368, min: 1.392856437922212, max: 34.41406140912264 PG1 time: 0.0905461311340332, mean: 13.000432968139648, sd : 3.201979160308838, min: 1.1816163063049316, max: 36.43503189086914
### Environment
* Windows 10
* Python 3.8.5 (anaconda)
* MSVC 2019
* RTX 2080Ti x2 + GTX 1070


1. Summary
Backend of CTnoise for Python and addCTnoise for Matlab are replaced with CUDA implementation.
Motivation
I bought new Matlab, but no money to buy Matlab Image Processing Toolbox nor Statictics Toolbox. ToT)