This is a known issue that I have already flagged with the cytoverse team. Hopefully support will be added for this soon.
Open rwbaer opened 2 years ago
This is a known issue that I have already flagged with the cytoverse team. Hopefully support will be added for this soon.
Thanks @DillonHammill. I have been trying to keep my workflow gatingSet and CytoExploreR-centric, but understanding all the things you take care of for us behind the scenes (and when I might be abusing your assumptions) can get intimidating. Your feedback is very helpful.
Describe the bug Transformation is apparently lost during save and load of gating set.
Expected behavior My expectation is that when I save a gatingSet with cyto_save() and the transform list has been specified by the 'trans = argument' that it will come back in as properly transformed data equivalent to what I created from raw .fcs files in CytoExplorer. The loaded data set should be usable in ggcyto().
Actual behavior Apparent bug. [1-works as expected] I load spill correct, and transform a gating set. I create some additional gates which I add to gating set and recompute(gsSamp), I then plot it in ggcyto() including the lines:
The plot axes are drawn, as expected, in original units.
[2- no longer works as expected] I then do a cyto_save(gsSamp, trans = TransList) followed by cyto_load(gsSamp) and plotting the very same plot no longer honors the axis_x_inverse_trans() and axis_x_inverse_trans() functions
Below are the graphs before and after the save/reload
Screenshots Expected graph (graph as drawn before cyto_save() and cyto_reload():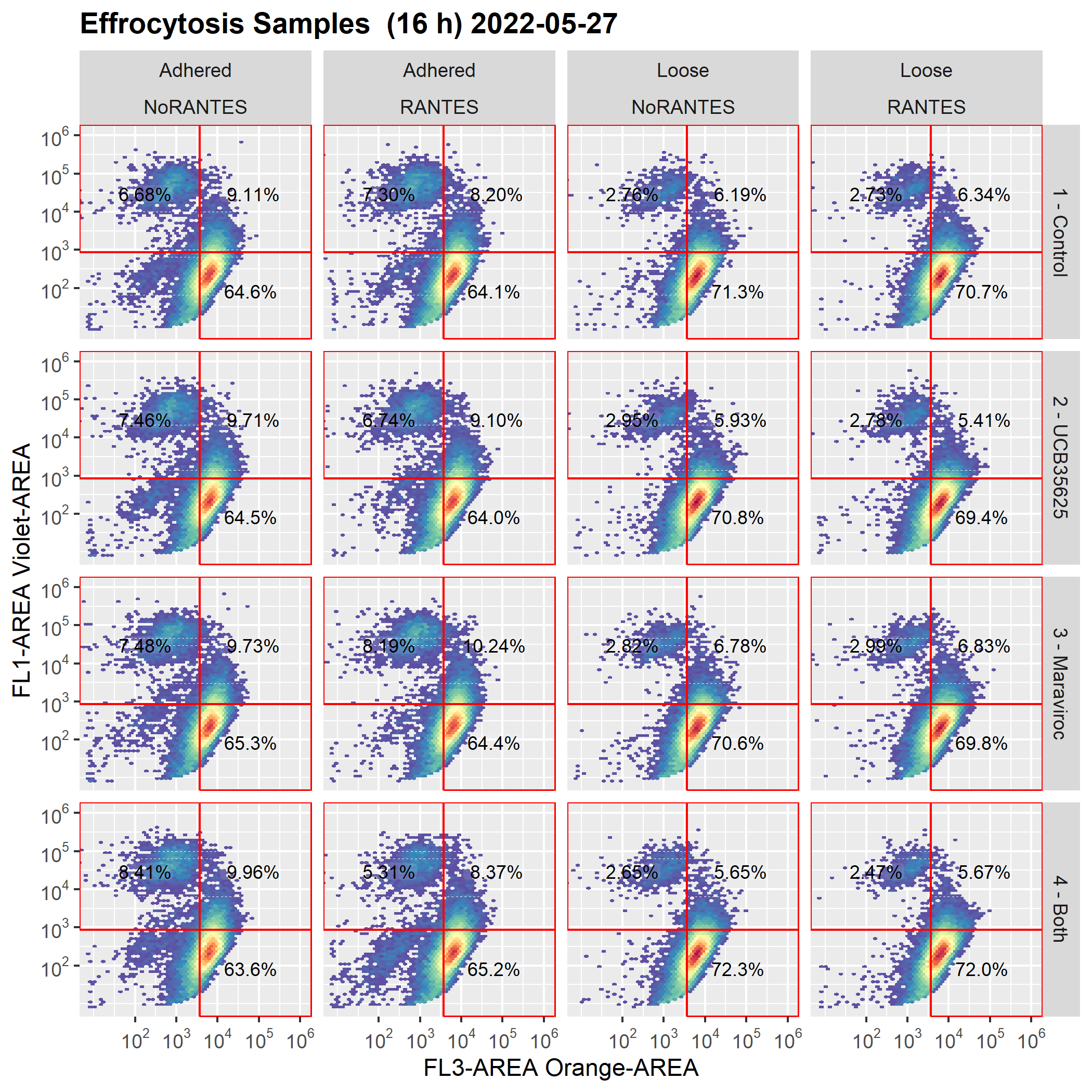
Graph after cyto_save() and cyto_load()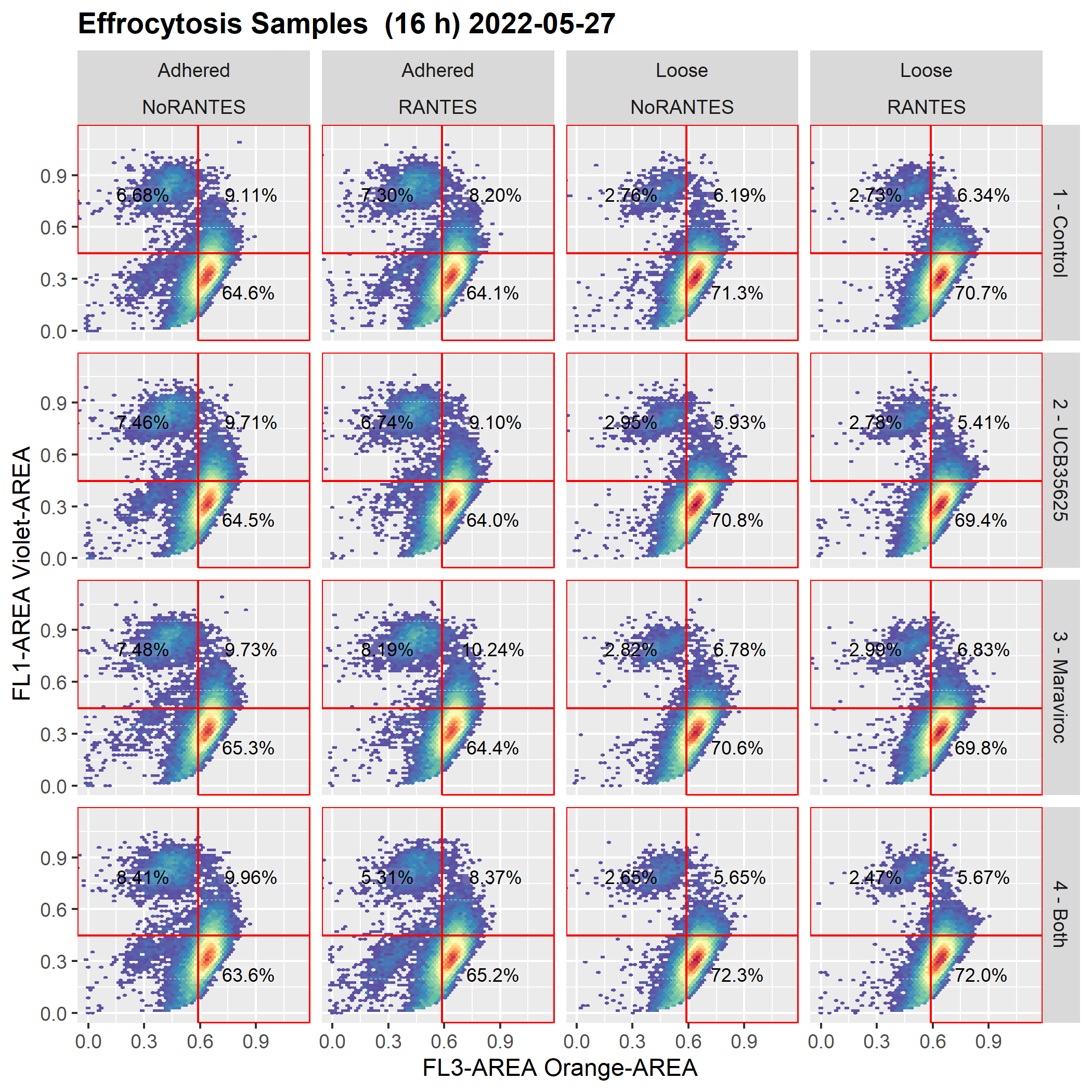
Additional context - more complete code for the repex