Another consideration is that gene level annotations can contain both processed transcript children and non coding children. The gene glyph could be extended to handle this. Currently it just displays a box for the non coding children, but it could be coded to display the "segments" glyph (and maybe color it differently)
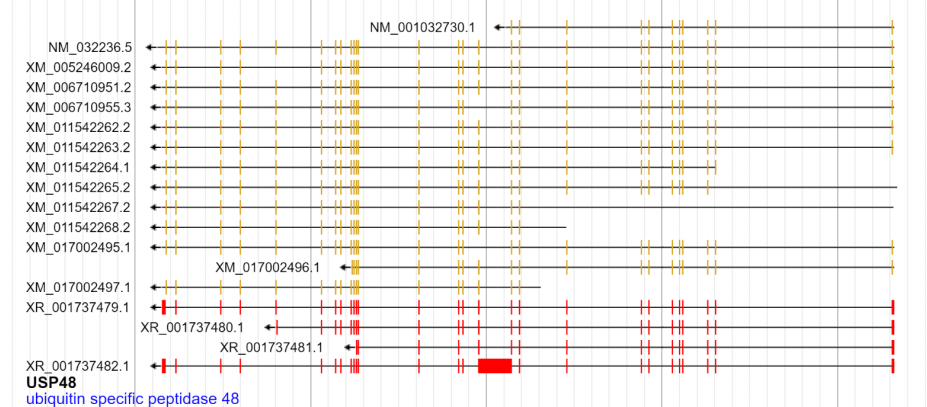
Some basic code for this here https://github.com/GMOD/jbrowse/tree/unprocessed_transcript_glyph
Weird corner case is that the style.color of the higher level feature is having trouble overriding the lower level box glyphs goldenrod
The issue discussed in #1075 highlighted that representing pseudogenes is a little tricky
Especially with a track that has both genes and pseudogenes, it would be good to dispatch to the Gene glyph for gene feature types and a Psuedogene glyph for pseudogene features in CanvasFeatures, because pseudogenes don't share the same structure of having CDS for example so their structure is not captured well