Possible dupe https://github.com/GMOD/jbrowse/issues/490
Hiding track labels is one way this has been "solved" with HideTrackLabels but I agree it would be better to just have at least the option to move the features out of the way (kind of what I wanted to say on that issue with "reconfiguring the UI")
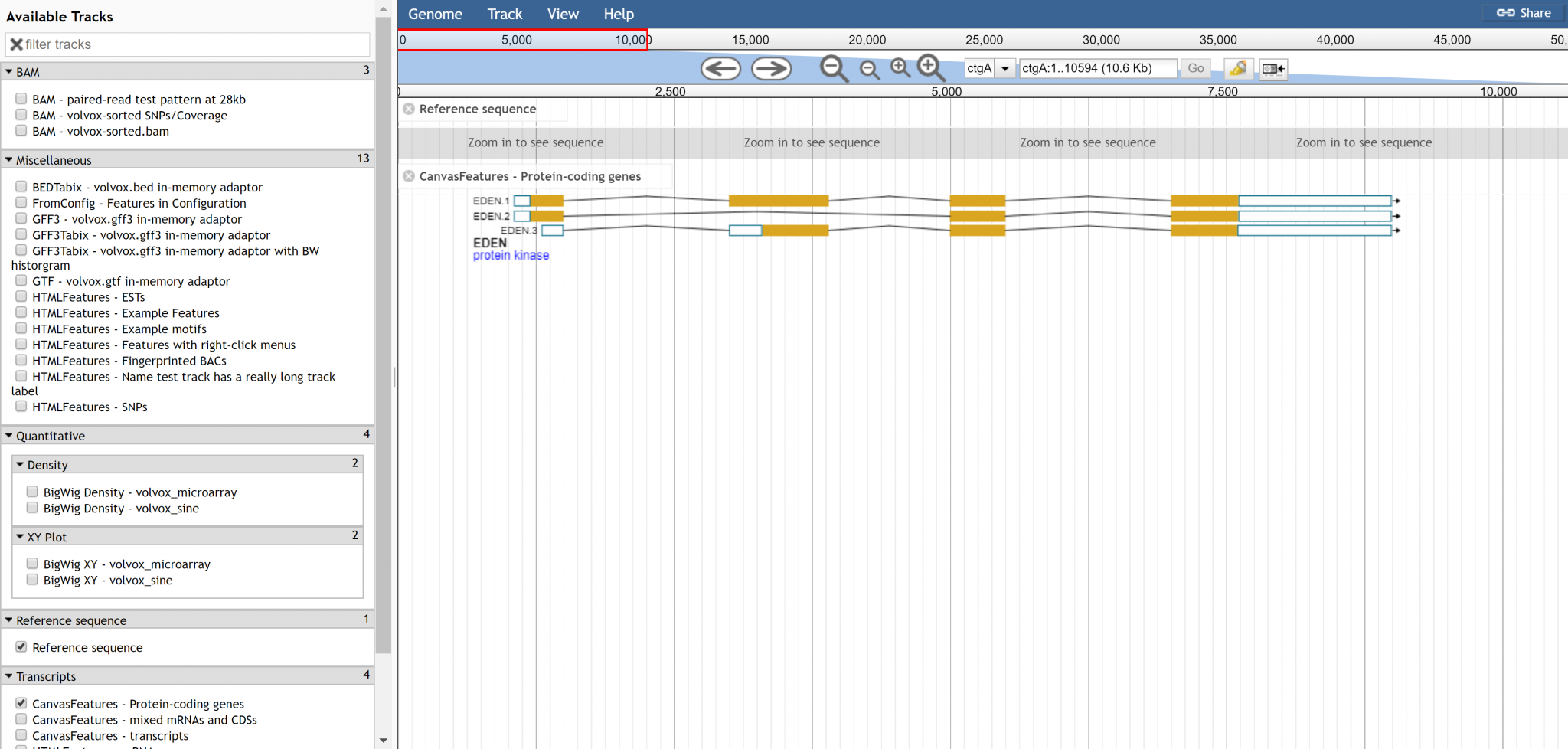

We like having track labels displayed, but could an option exist to offset the label from the features in the track? We often have very deep tracks so the additional depth from having the label Y-offset from the topmost feature is a very small price to pay compared to not being able to read a feature label or click on a feature without scrolling over horizontally to clear the track label.