Hello,
I am working with Barley which have very large chromosomes. Could you please suggest a way in which I could visualize the alignments in JBrowse and still bypass the issue with indices.
Thank you.
Closed keiranmraine closed 6 years ago
Hello,
I am working with Barley which have very large chromosomes. Could you please suggest a way in which I could visualize the alignments in JBrowse and still bypass the issue with indices.
Thank you.
FYI: https://samtools.github.io/hts-specs/SAMv1.pdf
5.3 C source code for computing bin number and overlapping bins
The following functions compute bin numbers and overlaps for a BAI-style binning scheme with 6 levels and
a minimum bin size of 214 bp. See the CSI specification for generalisations of these functions designed for
binning schemes with arbitrary depth and sizes.
/* calculate bin given an alignment covering [beg,end) (zero-based, half-closed-half-open) */
int reg2bin(int beg, int end)
{
--end;
if (beg>>14 == end>>14) return ((1<<15)-1)/7 + (beg>>14);
if (beg>>17 == end>>17) return ((1<<12)-1)/7 + (beg>>17);
if (beg>>20 == end>>20) return ((1<<9)-1)/7 + (beg>>20);
if (beg>>23 == end>>23) return ((1<<6)-1)/7 + (beg>>23);
if (beg>>26 == end>>26) return ((1<<3)-1)/7 + (beg>>26);
return 0;
}
/* calculate the list of bins that may overlap with region [beg,end) (zero-based) */
#define MAX_BIN (((1<<18)-1)/7)
int reg2bins(int beg, int end, uint16_t list[MAX_BIN])
{
int i = 0, k;
--end;
list[i++] = 0;
for (k = 1 + (beg>>26); k <= 1 + (end>>26); ++k) list[i++] = k;
for (k = 9 + (beg>>23); k <= 9 + (end>>23); ++k) list[i++] = k;
for (k = 73 + (beg>>20); k <= 73 + (end>>20); ++k) list[i++] = k;
for (k = 585 + (beg>>17); k <= 585 + (end>>17); ++k) list[i++] = k;
for (k = 4681 + (beg>>14); k <= 4681 + (end>>14); ++k) list[i++] = k;
return i;
}FYI, csi also applies to files that have traditionally used tabix indexing *.tbi:
$ tabix -h
...
Indexing Options:
...
-C, --csi generate CSI index for VCF (default is TBI)Hello
Large VCF files need also to be indexed using CSI index, so JBrowse cannot handle them right now.
Began some basic csi (for vcf currently) parsing here https://github.com/GMOD/jbrowse/tree/csi_index
Woo! tested and it displays data in super big coordinates that tabix tbi can't index (when chromosome over a gigabase in length)
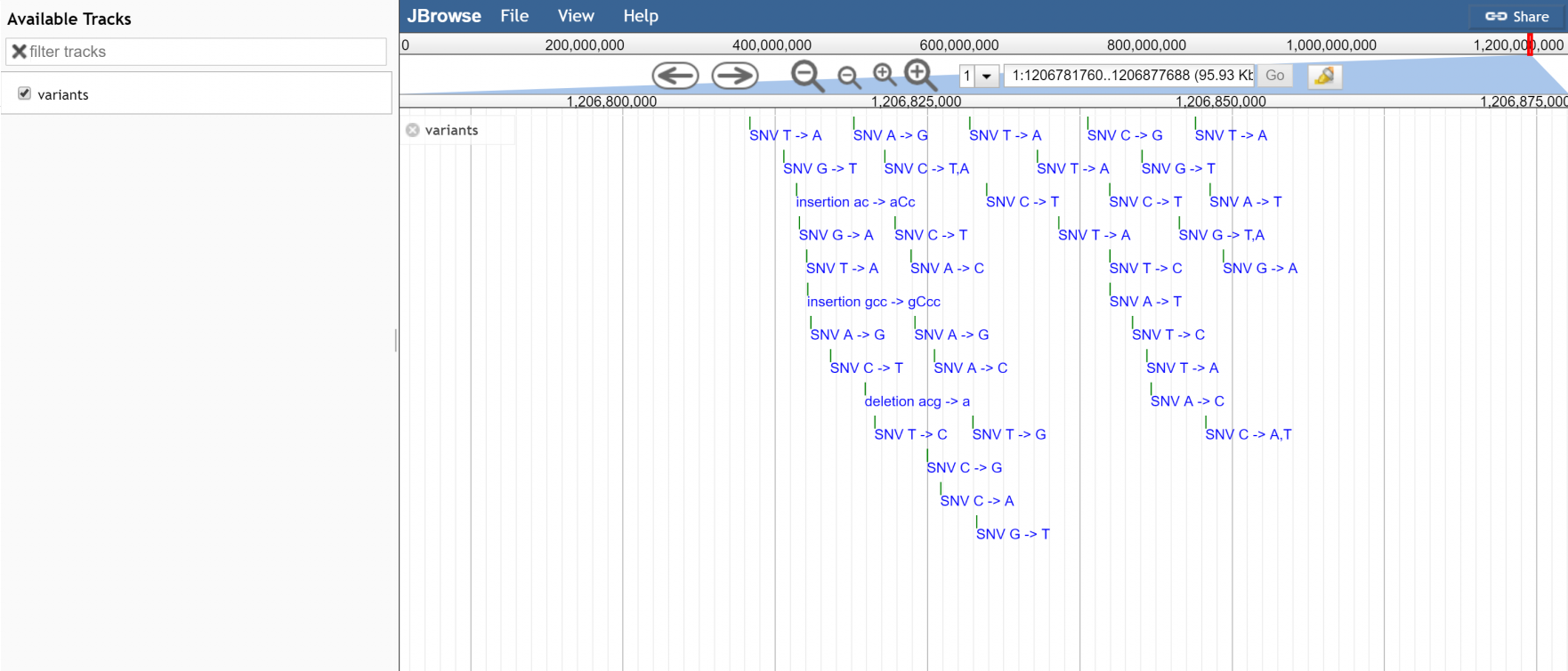
Got CSI working for BAM now also :) woo
Oh man...I almost wet myself with excitement! I want to test this out ASAP with wheat! :)
1 happy man at this prospect!
... do I dare say that they are currently discussing/adding *.sbi indexing:
http://github.com/samtools/hts-specs/pull/321
(will help solve the "guessing" about chunks)
Oh wow haha. Is that an official solution to "bam index index"?
I'm pretty sure that it doesn't but it's worth being aware that
*.csiindexes will replace*.baieventually, even for*.bamfiles.https://github.com/samtools/hts-specs/issues/240#issuecomment-328872226
I'm not aware of any progress on migration htslib based parsing of
bam/cram.