Verifying the computations
You can follow along by running the following script:
# ---- load-packages -----------------------------------------------------------
library(tidyverse)
# ---- load-data -------------------------------------------------------------
variable_names <- c("study_name","process_a", "process_b", "subgroup", "n", "est", "se")
proto <- list(
row01 = c("OCTO" ,"PEF" ,"Clock" ,"Men" ,138 , 0.27 ,0.14),
row02 = c("OCTO" ,"PEF" ,"Clock" ,"Women" ,275 , 0.24 ,0.11),
row03 = c("MAP" ,"FEV1" ,"Ideas" ,"Men" ,321 ,-0.02 ,0.08),
row04 = c("MAP" ,"FEV1" ,"Ideas" ,"Women" ,935 , 0.19 ,0.06),
row05 = c("EAS" ,"PEF" ,"MMSE" ,"Men" ,324 , 0.31 ,0.16),
row06 = c("EAS" ,"PEF" ,"MMSE" ,"Women" ,545 , 0.18 ,0.10),
row07 = c("MAP" ,"FEV1" ,"MMSE" ,"Men" ,321 , 0.04 ,0.08),
row08 = c("MAP" ,"FEV1" ,"MMSE" ,"Women" ,935 , 0.02 ,0.06),
row09 = c("NAS" ,"FEV1" ,"MMSE" ,"Men" ,1131 , 0.20 ,0.07),
row10 = c("OCTO" ,"PEF" ,"MMSE" ,"Men" ,140 , 0.66 ,0.14),
row11 = c("OCTO" ,"PEF" ,"MMSE" ,"Women" ,276 , 0.10 ,0.13),
row12 = c("SATSA" ,"FEV1" ,"MMSE" ,"Men" ,299 , 0.16 ,0.12),
row13 = c("SATSA" ,"FEV1" ,"MMSE" ,"Women" ,411 ,-0.07 ,0.18)
) %>%
dplyr::bind_rows() %>%
t() %>%
tibble::as_tibble()
names(proto) <- variable_names
proto[,c("n","est","se")] <- sapply(proto[,c("n","est","se")], as.numeric)This will recreate the data from the spreadsheet for the domain "Mental Status". This inputs all the data needed for analysis.
Question
I do not see where the spreadsheet utilized sample size. Are computations of the effect sized corrected for sample size? If yes, where?
Moving on
Operating on the object proto that contains estimates for the selected domain we compute the values in the green section:
# ---- tweak-data --------------------------------------------------------------
# green section: compute CI for observed estimates
ds1 <- proto %>%
# compute lower and upper limit of the 95% confidence (green section) for units
dplyr::mutate(
s = est, # Value of the estimate
w = log( (1 + s)/(1 - s) )/2, # ? w
u = (w * se)/ est, # ? u - Value for standard error?
ab = u * 1.96, # ? ab
aa = w + ab, # ? aa
z = w - ab, # ? z
y = ( (exp(2*aa)-1) / (exp(2*aa)+1) ) - s, # ? y
x = s - ( (exp(2*z)-1) / (exp(2*z)+1) ) , # ? x
lo = -(x - s), # low 95% CI
hi = y + s, # high 95% CI
ac = abs( w/(u^2) ), # ? ac
ae = u^-2, # ? ae | same as ag
ai = (z / 1.96)^2*ae, # ? Q
aj = ae / sum(ae) * 100 # ? node size
)
ds1 %>% dplyr::select_(.dots = c(variable_names,"s","u","w", "x","y","z","aa","ab","lo","hi"))The output is better captured in a screenshot.
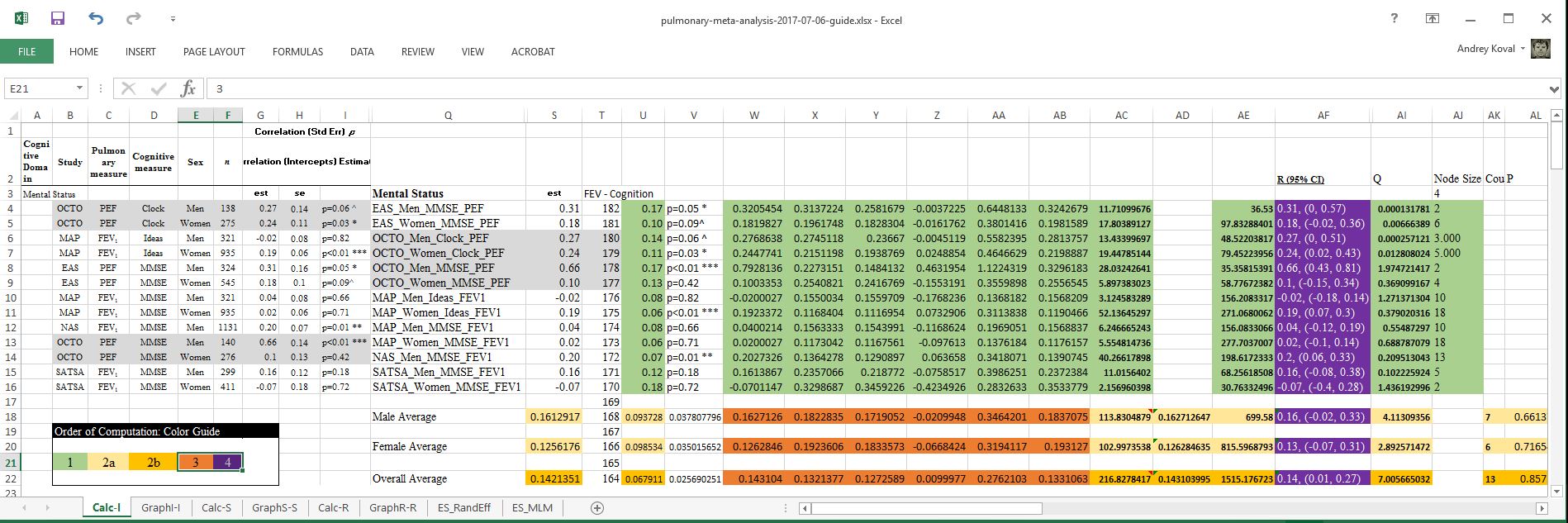
You can see that the values are identical to the values in the green section:
Annotating meta-analysis
In pursuit of creating a transparent record of all computations undertaken during the implementation of meta-analysis and creation of the forest plots (see pulmonary-meta-analysis-2017-06-20.xlsx ), we have created a translation of the computations to be implemented in R and demonstrated this process with the R script ./sandbox/meta-analysis-demo.R and the excel companion guide pulmonary-meta-analysis-2017-07-06-guide.xlsx. The code and images to follow refer to contents of these files.
In the original spreadsheet, I have identified the cells that are actively engaged in producing the forest plot, located in the shaded area in the screenshot below
The forest plot requires the following values: estimate, confidence intervals, and node size. The tricky part is to compute these values for subgroup and overall averages, which requires additional computaion.
After cosmetic editing, we can tidy up the spreadsheet into the form that would easier to handle during annotation.
Finally, to demonstrate how the computations are carried out in R starting from the same data source, specific cells were colored to illustrate the sequence in which computations took place. The number of the sections (e.g. "1", "2a", "2b", "3") are referenced in the script ./sandbox/meta-analysis-demo.R . which details each set of operations.
This will be the main graphs referenced in this discussion.