Codecov Report
Merging #5 into master will not change coverage. The diff coverage is
100%.
@@ Coverage Diff @@
## master #5 +/- ##
=====================================
Coverage 100% 100%
=====================================
Files 3 5 +2
Lines 210 281 +71
=====================================
+ Hits 210 281 +71| Impacted Files | Coverage Δ | |
|---|---|---|
| src/core.jl | 100% <ø> (ø) |
:arrow_up: |
| src/region_growing.jl | 100% <100%> (ø) |
:arrow_up: |
| src/ImageSegmentation.jl | 100% <100%> (ø) |
|
| src/fast_scanning.jl | 100% <100%> (ø) |
Continue to review full report at Codecov.
Legend - Click here to learn more
Δ = absolute <relative> (impact),ø = not affected,? = missing dataPowered by Codecov. Last update 0d8720a...22e05e5. Read the comment docs.

An implementation of the fast scanning algorithm as mentioned in this paper. An extension of this algorithm has also been implemented for working with higher dimensional images. A few features like removal of small segments and use of adaptive threshold will be progressively added. The performance of this algorithm for 2-d images is fantastic. After addition of the above-mentioned features, I will add the docstring and tests.
Here are a demo:
Original: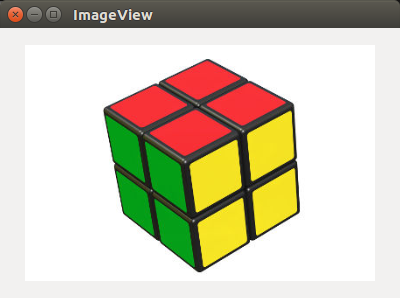
After segmentation and coloring the larger segments (except background):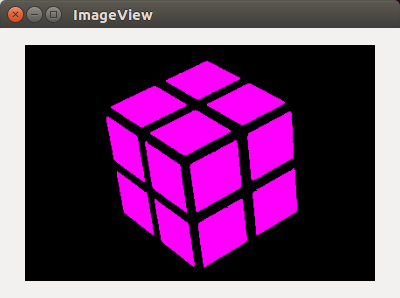
Benchmark results: