@tilmantroester in this case the mask (and the fields) you're using are complete non-band-limited (i.e. the power spectrum of the mask is constant at high ell), so the fact that namaster only computes mode-coupling up to a given ell_max coupled with the numerical errors of healpix means that the resulting MCM is probably inaccurate.
Maybe try using a nicer mask (e.g. a smoothed version of this one)?


I have an issue where namaster seems to fail to return unbiased estimates of the power spectrum if a weight map is being used as the mask. I don't know if that's a fundamental issue (doubtful, since support for weight maps is stated in a couple of places) or I messed up somewhere (entirely possible but the same code works for binary masks). Apodisation doesn't seem to help.
The setup is as follows (full code at the end):
nmt_workspace.decouple_cell(nmt_workspace.couple_cell(Cl_theory))For full-sky (top panel) and binary mask (bottom panel, just the circular footprint), this works as expected: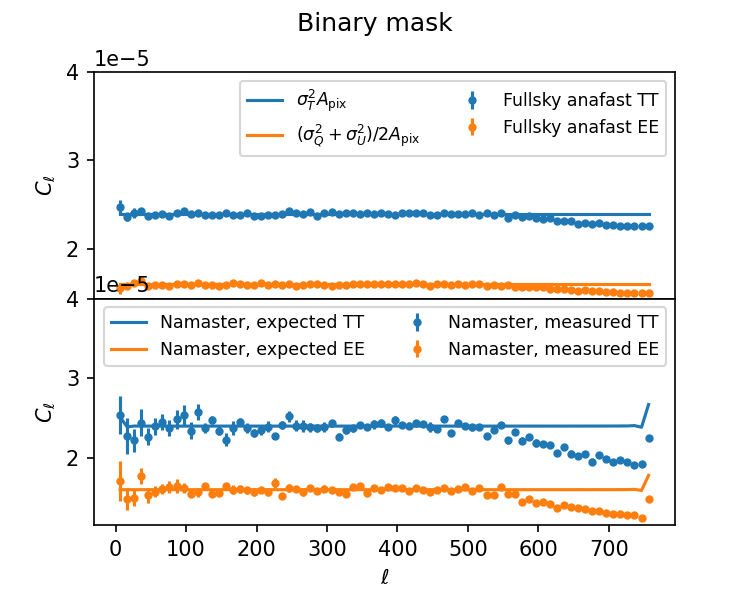
When using the weight map, the measured, decoupled Cls don't agree with the analytic values anymore: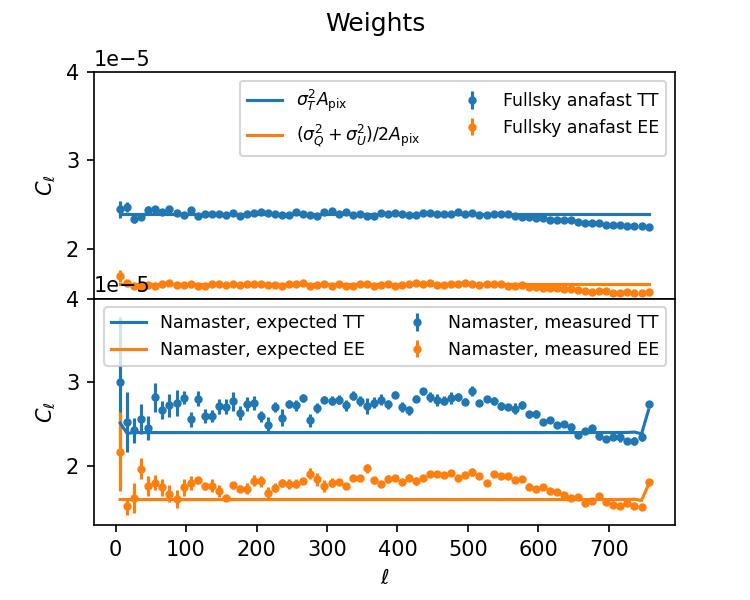
Code: