Closed jkrumbiegel closed 2 weeks ago
2D contour plots would be VertexSurfaceLike by this logic (important for the isoband PR that I want to merge soon as well)
That is a very important issue that I think should be addressed more seriously at the level of mesh types. I've been working on Meshes.jl where I have this in mind as well. Currently the two available meshes CartesianGrid and UnstructuredMesh are VextexCentered with the first just a lazy construct on top of ranges. For the CellCentered equivalent, I am planning to add a ImageGrid or figure out a clever design that avoids code duplication.
Rectilinear and curvilinear meshes, also deserve their own types, and can be written compactly on top of primitives and other types of geometries.
Let's maybe separate mesh representation issues from the issue here, which is just about drawing these things correctly. Conversions from special types can come later I think, because the underlying representation on the GPU will be the same for all of these.
I am not familiar with the traits that are implemented in the Makie.jl stack, but it would certainly be useful to have them aligned with a collection of mesh types. I agree that the issue can be brainstormed regardless of the mesh representation.
GLMakie currently just draw one textured Rectangle for both image and mesh. For surfaces we use either a Grid2D atm, which is essentially like two ranges, or two Vectors or two matrices as x/y inputs. The vertices, normals and colors are then computed on the gpu. I think the same would be fine for heatmaps.
To me the difference between VertexSurfaceLike and FaceSurfaceLike is just the difference between image and heatmap. An image is one object, it has a starting position, width and height and that's it. A heatmap is a collection of cells at some positions, each with a color. To me that implies that each cell would represent one point, i.e. that each cell is face centered.
I would suggest that image implements methods like
image(img::Matrix)
image(origin::Point, widths::Vec, img)
image(origin::Point, endpoint::Point, img)
image([x_min, x_max], [y_min, y_max], img)
image([x_min, x_max], [y_min, y_max], [z_min, z_max], img)
image(r::Rect, img)corresponding to VertexSurfaceLike case and heatmap has some changes internally to implement (just) the FaceSurfaceLike case. I would also suggest dropping Matrix inputs for x and y, because in the face centered case it would require computing what these (irregular) faces look like. That seems like a lot of work and also something that would generate a mesh on the cpu which, well, should probably be a mesh-plot at that point.
I thought the difference between image and heatmap was that an image comes with color data per pixel, and not numerical values that get interpreted via colormap. But I agree that intuitively, one thinks more of the default rectangular image.
In your view, should pixel [1, 1] of an image starting at x=1 be centered at 1 or 1.5? That's the difference between specifying the boundaries or specifying the boundary cell centers. I'm not sure if it's good if heatmaps and images differ in that regard. I think I favor the version where pixel centers are what you specify, because people often want to mark positions in images, and that makes more sense if you can just count the pixels.
Matlab also does it that way:

fwiw the cellsurfacelike behaviour you are suggesting is the same as that for 2d bar plots (where you supplement the mid point) and vertex-surfacelike the one for histograms (where you supply the bin edges). Bonus info: Plots offers the double api for bar plot where you can supply either the mid points OR n+1 limits between bars (which I think is not a clean design btw).
I thought the difference between image and heatmap was that an image comes with color data per pixel, and not numerical values that get interpreted via colormap. But I agree that intuitively, one thinks more of the default rectangular image.
Currently image and heatmap are more or less the same thing with different presets. You can call image(5:15, 1:10, rand(10, 10), colormap=:viridis, interpolate=false) to get what heatmap(5:15, 1:10, rand(10, 10)) creates. The other way doesn't quite work but it only requires one or two lines in GLMakie to be changed.
In your view, should pixel [1, 1] of an image starting at x=1 be centered at 1 or 1.5? That's the difference between specifying the boundaries or specifying the boundary cell centers. I'm not sure if it's good if heatmaps and images differ in that regard. I think I favor the version where pixel centers are what you specify, because people often want to mark positions in images, and that makes more sense if you can just count the pixels.
Having an image pixel-centered is weird to me, but I do see the point. Probably best to keep things very adjustable then.
Bonus info: Plots offers the double api for bar plot where you can supply either the mid points OR n+1 limits between bars (which I think is not a clean design btw).
I was thinking about that too. If we were to allow both options this would be a way to handle it. Though it would be unclear how to handle 2-element arrays. The other option would be to have an attribute to specify where cell centers are, which sounds better to me.
Maybe then we should have
image([xs, ys], img_or_mat[, align=:outside, interpolate=true, colormap=[:black, :white]])
heatmap([xs, ys], img_or_mat[, align=:center, interpolate=false, colormap=:viridis])with xs and ys being Intervals, two element vectors/ranges (tuples?) (min, max) or N element vectors/ranges (matching N cells)? Should we also support image/heatmap(origin::Point, widths::Vec, ...)?
Bonus info: Plots offers the double api for bar plot where you can supply either the mid points OR n+1 limits between bars (which I think is not a clean design btw).
I think that might be a bit brittle, and it's better to be explicit about the choice of centers vs. edges.
I would favor defaulting to centers (as least for heatmaps, but probably also for images). The error message when passing n+1 points as centers can give a hint about the relevant keyword to pass edges, or even suggest using StatsBase.midpoints to get the actual centers.
Also, here my recipe for centered heatmaps (just for working around this issue in the meantime, not as a suggestion for implementation):
cheatmap_range(r::AbstractRange) = let d=step(r)/2
range( first(r)-d, last(r)+d; length=length(r) )
end
cheatmap_data( x, y, z::AbstractMatrix ) = z
function cheatmap_data( x, y, z::Function )
dx, dy = step(x)/2, step(y)/2
fx, lx = extrema(x)
fy, ly = extrema(y)
fx0, lx0 = fx-dx, lx+dx
fy0, ly0 = fy-dy, ly+dy
(i,j) -> z( fx + (lx-fx)*(i-fx0)/(lx0-fx0),
fy + (ly-fy)*(j-fy0)/(ly0-fy0) )
end
@recipe(scene->Theme(), CHeatMap, x, y, z)
function AbstractPlotting.plot!(p::CHeatMap)
x,y,z = p[:x], p[:y], p[:z]
heatmap!( p, lift(cheatmap_range,x), lift(cheatmap_range,y),
lift(cheatmap_data,x,y,z); p.attributes... )
p
endFirst of, thanks all involved for looking into these! I'd like to help this move forward however I can, so I'll chime in to synthesize my understanding of the issue and hopefully someone can draw on it, correct my mistakes, and maybe that helps the devs to decide how to implement some changes! 😄
Currently, there are 2 truly flat 1 SurfaceLike plot types:
heatmap, a 2D array of cells, where each cell's color is interpolated from a colormap.image, a 2D array of colors.My understanding is that this issue is driven by the syntactic sugar we are accustomed to for these types of plots. When I run one of
image(rand(5,3))
heatmap(rand(5,3))
heatmap(1:5, 1:3, rand(5,3))I expect the plotting package to figure out the location and shape of the cells/pixels under the hood, and this issue is about doing just that, deciding what to do with different incomplete inputs.
About the 2-element vector confusion, I propose appending @jkrumbiegel's original post table with one row for x or y given as a Tuple, so for (1,10) to be interpreted as 1..10 is in the table. That would also modify @ffreyer's suggestion to replace the 2-element vectors with tuples, specifically:
image((x_min, x_max), (y_min, y_max), img)
image((x_min, x_max), (y_min, y_max), (z_min, z_max), img)Some of the issues referenced above request the feature to expand these to allow for less regular meshes. For example, one might want a heatmap where cells are in an irregularly-spaced rectilinear grid, in a curvilinear grid, or an otherwise generally unstructured mesh. I feel like these apply only to heatmap here, but some of the issues mentioned contour plots as well. Either way, I'll assume we are talking about heatmap hereon.
FWIW, my uneducated feeling is that @juliohm is right that having this discussion at the mesh level is the best approach to clearly delineate the different plot types/behaviors on that topic. But maybe I am missing something.
So, for heatmap, building on @jkrumbiegel's table, I would add
# if z is a m×n 2D array
heatmap(x::Tuple, y::Tuple, z::Array{<:Number,2}) = heatmap(x[1]..x[2], y[1]..y[2], z) # so same as ClosedInterval case
function heatmap(x::Vector, y::Vector, z::Array{<:Number,2}) # if `x` and `y` are vectors
if (length(x), length(y)) == size(z)
# x and y are cell centers
elseif (length(x), length(y)) == size(z) .+ 1
# x and y are cell edges
else
error("Sizes must be...")
end
end
heatmap(x::Array{<:Number,2}, y::Array{<:Number,2}, z::Array{<:Number,2}) = ... # regular mesh (can be cartesian, rectilinear, or curvilinear)
heatmap(x::Vector, y::Vector, z::Vector) # unstructured meshHopefully someone finds this useful 🙏
1: By truly flat I mean that is not thought of as embedded in a 3D space. For instance, I think surface might need its own separate category because surface suggests, to me, a 2D sheet in a 3D space. Of course one can think of the projection of a surface on a flat plane, but I don't think that's what this issue is about.
Okay I have a new idea about this. Our base issue is that heatmaps / images have an unusual behavior in that they go edge-to-edge and are not centered on their cells. Their colors are given per cell and not at the vertices, unlike typical meshes.
Images are actually not different from heatmaps, only that they are more constrained. For the implementation it does not really matter.
I don't really like having a "do we have n or n+1 numbers in our vectors" rule to decide between edge-based and center-based, as then from the call signature the behavior is not really clear anymore.
I would propose adding a simple type Edges which wraps anything that's supposed to be edges, and keep the center syntax as the default because it's the most common scenario by far.
In the backend, everything would work via Edges. We could assert that they're sorted or whatever we want in the Edges constructor to assure correct behavior.
Usage would look like this:
heatmap(rand(3, 3)) # centered on 1 to centered on 3 in both dimensions
heatmap(1:3, 1:3, rand(3, 3)) # same
heatmap(1..3, 1..3, rand(3, 3)) # same
heatmap([1, 2, 4], [1, 3, 5], rand(3, 3)) # unequal cell sizes, still giving the centers
heatmap(Edges(1..3), Edges(1..3), rand(3, 3)) # now the edges go from 1 to 3, not the centers
heatmap(Edges(1..3, 1..3), rand(3, 3)) # same, maybe more convenient with only one `Edges`
heatmap(Edges([1, 2, 4, 5], [2, 5, 6, 7]), rand(3, 3)) # unequal cell sizesThe "normal" signatures would be immediately converted to Edges and the backend would only deal with Edges
[EDIT: I just wanted to note that I started this comment a while ago and just finished it now without seeing @jkrumbiegel's latest comment. So sorry if I just added some noise 😓]
[EDIT 2: Adding that I like (love?) @jkrumbiegel's idea above]
I would really like to see this move forward, so I'm pushing with another supportive comment. Again, I hope this is somewhat useful.
My understanding at this point is that we have 4 types of grids/meshes in the end:

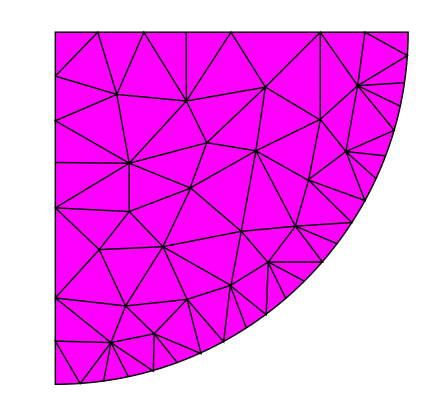
Right now, only cartesian grids are supported, except surface, which can deal with rectilinear grids (IIRC, although I am most likely wrong about this). Proof-of-concepts in the referenced issues https://github.com/JuliaPlots/CairoMakie.jl/pull/111 and https://github.com/JuliaPlots/GLMakie.jl/pull/137 were aimed at allowing rectilinear grids for heatmap.
I do think that, in the long run, being able to plot any of image, surface, heatmap, or contour, contourf, and so on, on an unstructured mesh, would be invaluable to efforts such as GeoMakie. However, maybe it is too ambitious to try and aim from the start for the holy grail? What if, instead, we laid down a plan to extend Makie's capabilities one step at a time, by first tackling the rectilinear case, and then the more complex cases?
So what is needed for the rectilinear case? Well, it seems that the backends only need to be able to deal with the more general rectilinear meshes. So IMHO the default should be that at the AbstractPlotting level, plot types returned are actually defining the edges of the cells. So I would assume we could implement the following rules (which is slightly different take from what @jkrumbiegel — who is much more knowledgeable than me — suggested):
x or y is an Interval or a Tuple then it gets turned into a range of the edges, e.g., x=(-10,10) gets converted to range(x[1], x[2], length=size(z,1)+1) (although maybe I'm confused about which dimension of z should correspond to x, but that's another debate 😅)x is an AbstractVector (which includes ranges and vectors) then its length is checked against z:
x values are the midpoints, so we infer the edges from xx is that of z + 1, then x values are already the edges and we're doneI'm not saying that the end product (at the backend level), everything should be of the rectilinear, cells-defined-by-edges type. My uneducated guess is that depending on the type of plot returned by AbstractPlotting, e.g., a heatmap of which the x and y values are ranges, the backend could then dispatch to a simpler (more economical) plot type to display or print, inferring from x::AbstractRange and y::AbstractRange that the cells are regularly spaced (maybe using a scaled pixelated image in this particular example).
I'm personally usually for doing it "right" immediately, so if there's great need for the more complex scenarios we should incorporate those as well I guess. In the unstructured example we don't even have a grid-like structure anymore though, does that still make sense for heatmap/image? That looks very mesh-y to me. Or is this still about coloring each face separately instead of the vertices?
I agree that it's always better to get it right soon. Here are some real-world examples:
this is taken from a paper about the MPAS Ocean model, which has a good-looking Voronoi-type of hexagonal mesh of which the resolution varies depending on location:

Another example from the CESM model, which uses a lat-lon grid with 3 poles (below you can see the 2 north poles), which curves the grid substantially in some places:

How cool would it be to be able to make heatmaps on these meshes?
Anyway, back to what you were saying, I think that it makes sense to have, e.g., heatmap, color each face separately, as it seems like the most generic approach. (And my impression is that it's probably a requirement for curvilinear grids anyway, even though they are structured.)
I think plots like those would fit more into mesh, but I believe we can't do face-colored meshes at the moment. We can do vertex-colored meshes though, which is essentially the same but with interpolation. (I've done something along those lines here but I don't know if it still works)
Also, how would we differentiate a curvelinear heatmap from normal ones? If there's no clear way to differentiate them, then curvelinear heatmaps should probably be their own separate thing. (Having different plot types for these might also be helpful in GLMakie. A curvelinear heatmap requires a different shader and maybe also a different primitive, I think.)
I'd suggest we get rectlinear heatmaps done first and leave the other fancy cases for later. I'm not a big user of heatmaps so I don't have a strong opinion on what the front end of this should look like. I'm fine with doing the Edges thing. What would we do with image in this case? Do you also want this to default to a centered view, with Edges being the special case?
Is there a level or two of abstraction between "rectilinear" and "unstructured", for Euclidean uniform tilings

and more general uniform tilings?

https://github.com/JuliaGeometry/VoronoiCells.jl might be useful for unstructured tiling?
jkrumbiegel wrote on jan 29:
Usage would look like this:
heatmap(rand(3, 3)) # centered on 1 to centered on 3 in both dimensions
heatmap(1:3, 1:3, rand(3, 3)) # same
heatmap(1..3, 1..3, rand(3, 3)) # same
heatmap([1, 2, 4], [1, 3, 5], rand(3, 3)) # unequal cell sizes, still giving the centers
heatmap(Edges(1..3), Edges(1..3), rand(3, 3)) # now the edges go from 1 to 3, not the centers
heatmap(Edges(1..3, 1..3), rand(3, 3)) # same, maybe more convenient with only one `Edges`
heatmap(Edges([1, 2, 4, 5], [2, 5, 6, 7]), rand(3, 3)) # unequal cell sizesI took this as an exercise.
GLMakie.activate!()
using Makie: IntervalSets
theme = Theme(fontsize = 12, colormap = :gist_earth, resolution = (500,300) )
set_theme!(theme)Benchmark using Makie v0.13.11:
@btime heatmap(rand(3, 3))16.292 ms (152297 allocations: 6.02 MiB)

my code:
@btime myheatmap(rand(3, 3)) # same (but note the use of natural numbers)7.594 ms (45968 allocations: 2.58 MiB) # the compiler seems to like the code ...

@btime myheatmap(1:3, 1:3, rand(3, 3) ) # same (note the use of natural numbers)7.737 ms (45971 allocations: 2.58 MiB)

@btime myheatmap(1..3, 1..3, rand(3, 3)) # same (but now with reals rather than rational numbers)16.229 ms (152299 allocations: 6.03 MiB)

@btime myheatmap([1, 2, 0.5, 4], [1.1, 3.2, 5.3], rand(4, 3) ) # unequal cells of given size8.027 ms (46658 allocations: 2.61 MiB)

Next we turn to 'the Edges'. I implemented this by setting a keyword: center=false:
@btime myheatmap(1..3, 1..3, rand(3, 3); center=false) # now the edges go from 1 to 3, not the centers16.201 ms (151591 allocations: 5.87 MiB)

@btime myheatmap(1..3, 1..3, rand(3, 3); center=false) # now the edges go from 1 to 3, not the centers16.201 ms (151591 allocations: 5.87 MiB)

@btime myheatmap((1:3), (1:3), rand(3, 3); center=false) # same but faster7.845 ms (45967 allocations: 2.58 MiB)

@btime myheatmap([1.4, 2.3, 0.5, 4.2], [1.1, 3.2, 5.3], rand(4, 3); center=false) # unequal cells with respect to edges8.374 ms (49314 allocations: 3.01 MiB)

You guys are really giving Makie a push towards broader application, maybe my code contains some lines of inspiration:
(File was edited to include autoticks.)
using Makie: IntervalSets
_log10_characteristic(x) = Base.round(Int,Base.floor(log10(x)))
_log10_mantissa(x) = Base.log10(x)-Base.floor(Base.log10(x))
function _autostep(x::Int)
m = _log10_mantissa(x)
p = _log10_characteristic(x)
v = 10^m
d = v > 7.9 ? 2.0 : v > 3.9 ? 1.0 : v > 1.49 ? 0.5 : 0.2
return max(1,round(Int, d *= 10^p))
end
function _autoticks(x)
n = length(x)
return [x[i] for i=_autostep(n):_autostep(n):n]
end
function to_center(interval, dim; center=true)
Δ = center ? 0.5 : 0
ρ = (interval.right - interval.left)/dim
return (Δ + interval.left + 0.5ρ)..(Δ + interval.right - 0.5ρ)
end
function to_ticks(segments::Vector{<:Real})
s = copy(segments)
o = [sum(s[1:i]) for i ∈ eachindex(s)]
popfirst!(s)
pop!(o)
return o .+ s .* 0.5f0
end
function myheatmap(σ)
tx = _autoticks(Base.OneTo(size(σ,1)))
ty = _autoticks(Base.OneTo(size(σ,2)))
return heatmap(σ, axis = (xticks=tx, yticks=ty,) )
end
function myheatmap(x::UnitRange{Int}, y::UnitRange{Int}, σ; center=true)
if !center
x = to_center(x[1]..x[end], size(σ,1); center=center)
y = to_center(y[1]..y[end], size(σ,2); center=center)
end
tx = _autoticks(Base.OneTo(size(σ,1)))
ty = _autoticks(Base.OneTo(size(σ,2)))
return Makie.heatmap(x, y, σ, axis = (xticks=tx, yticks=ty,) )
end
function myheatmap(x::Vector{<:Real}, y::Vector{<:Real}, σ; center=true)
length(x) == size(σ,1) || error("Error: number of x segments ($(length(x))) not equal to x dimension ($(size(σ,1)))")
length(y) == size(σ,2) || error("Error: number of y segments ($(length(y))) not equal to y dimension ($(size(σ,2)))")
prepend!(x,0)
prepend!(y,0)
if center
tx = (_autoticks(to_ticks(x)), string.(_autoticks(Base.OneTo(size(σ,1)))) )
ty = (_autoticks(to_ticks(y)), string.(_autoticks(Base.OneTo(size(σ,2)))) )
else
tx = _autoticks([sum(x[1:i]) for i ∈ eachindex(x)])
ty = _autoticks([sum(y[1:i]) for i ∈ eachindex(y)])
end
x = [sum(x[1:i]) for i ∈ eachindex(x)]
y = [sum(y[1:i]) for i ∈ eachindex(y)]
return Makie.heatmap(x, y, σ, axis = (xticks=tx, yticks=ty,) )
end
function myheatmap(x::IntervalSets.ClosedInterval{<:Real}, y::IntervalSets.ClosedInterval{<:Real}, σ; center=true)
if !center
x = to_center(x, size(σ,1); center=center)
y = to_center(y, size(σ,2); center=center)
end
return Makie.heatmap(x, y, σ)
endThis was more less implemented by #3106
We had multiple issues about nonlinear grids for heatmaps, but also about the bin centering of heatmaps and images (basically that pixel [1, 1] is not centered on point [1, 1]).
These discussions have mostly looked at implementing things in the backends, but I think it's an AbstractPlotting abstraction issue. Right now, there is a
SurfaceLiketrait which heatmap, image and surface use to convert their arguments. I think that this is incorrect, because they are different, even if only in a subtle way.The difference is that a surface colors the vertices of a grid, while image / heatmap colors the cells of a grid. I therefore suggest to remove
SurfaceLikeand instead addVertexSurfaceLikeandCellSurfaceLikeor something else to that effect. Maybe there are better words for this.Then the question is how the coordinates of these grids should be defined. Let's say our color data are always in the form of a matrix, then there are multiple different ways of defining the coordinates of vertices / cells.
Here's what I would suggest:
VertexSurfaceLikeCellSurfaceLike1.0..10.0Matrices would then just be extensions of the vector case.
One thing that this doesn't cover is a plot where the coordinates given are vertices, but the colors are for the cells. One could dispatch to that method by checking if the length of heatmap coordinates are n+1, but that might be a bit brittle?