Need probably many-to-many links between ExperimentalData and Protocol, and only once a fit is performed do you tie down exactly which were used? @MichaelClerx ?
Not sure what you mean here! Would an ExperimentalData be a single time series? (Or a single matrix, more abstractly)?
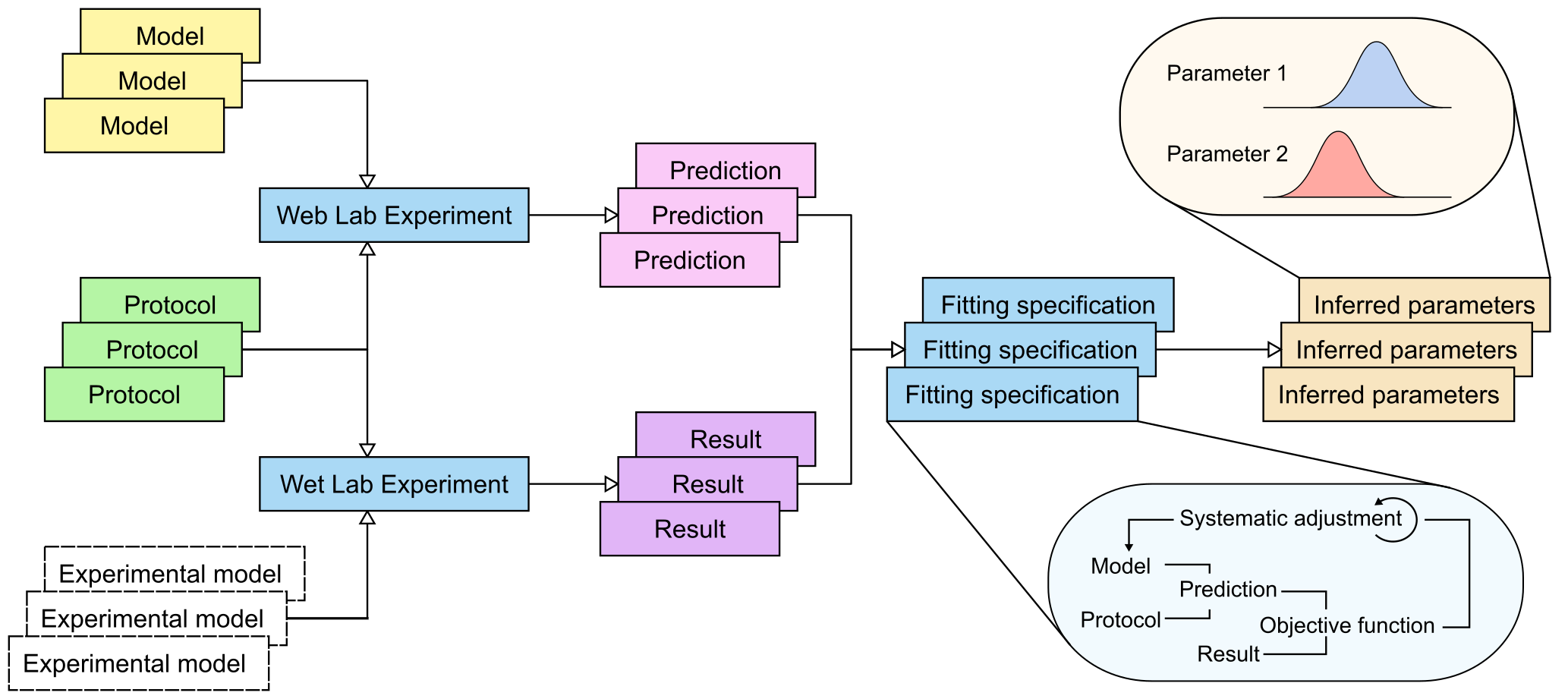
This may still get split into sub-issues...
[x] Figure out what is common with Experiment (cf #130) and put in a mixin class / common 'dataset' app (see also https://github.com/ModellingWebLab/project_issues/wiki/Workshop-notes-2018 and #203)
[x] Need probably many-to-many links between
ExperimentalDatasetandProtocol, and only once a fit is performed do you tie down exactly which were used? @MichaelClerx ?Add views to (these are probably almost identical to the entity views):
[x] Create an
ExperimentalDatasetmodel in its owndatasetsappEXPERIMENT_BASE, with a corresponding config setting giving the root path.UserCreatedModelMixin, VisibilityModelMixin, models.Modeland needs anameProtocol- this should be set on creation, e.g. from a drop-down list[x] Create views to (these are probably almost identical to the entity views):
datasets:archiveview like inexperiments/views.pydatasets:file_downloadview like inexperiments/views.py[ ] Add ability to compare data with predictions
dataset-linkinexperimentversion_detail.htmlandexperiment.js[ ] Add ability to compare data with data
ExperimentComparisonView