things to resolve and finish (if we decide to use Oxygen-generated documentation)
a) might want to copy over to a <xs:documentation> element, (and preserve xs:appinfo).
Plus: You would see the original xs:appinfo as nice XML in the block titled "Source". As is, the text content of xs:documentation is dumped to the screen.
Minus: you'd see the xs:appinfo content 3 times (converted at top, plus raw and original in source)
b) confirm this works for cdata sections. (cannot id a cdata section with xpath, but I think copy tranlates the examples within.)
c) script to run through all xsd files and dump in a temp folder (or some other)

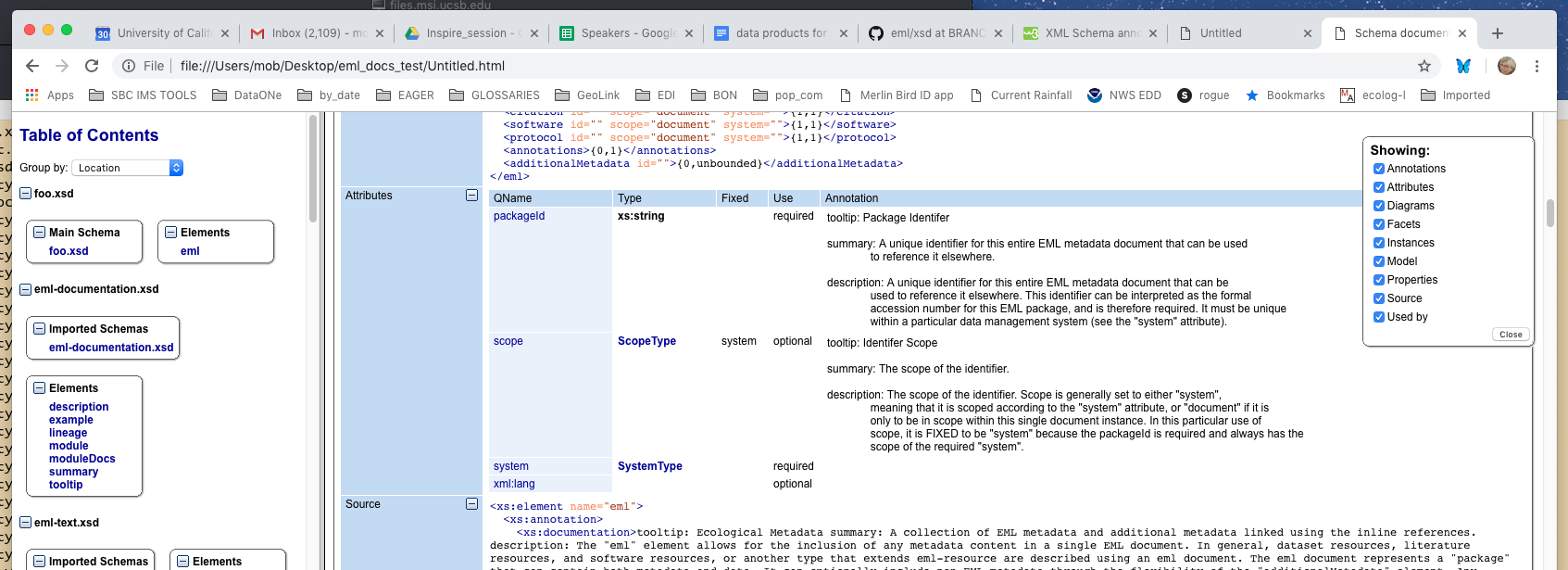
I did a bit of testing, to make use of the oxygen editor's documentation generation tool. The generation tool exports images and embedded documentation, for browsing.
Limitations: Oxygen only exports content of the xs:documentation element, EML uses xs:appinfo with docBook xml inside. So the appinfo content has to be moved to a documentation element, and it will only show plain text there. However,. the HTML rendering will respect newlines, etc, so we can use this to do a bit of formatting. Checked in stylesheet with these actions: a) copies an xsd file b) moves xs:appinfo content to a xs:documentation node, separated into functional paragraphs. c) uses each elements local-name as the label for that section.
process: xsltproc eml_appinfo2documentation.xsl ../xsd/eml.xsd > ../tmp/eml.xsd in OxygenXML editor: Tools > Generate Documentation > XML Schema Documentation
see attached screenshot