Dear @wangwei1619,
passive diffusion will only take place, if there is a difference in unbound concentration between the two spaces (plasma and interstitial). In MoBI you can select "create process rate parameter" and "plot parameter" to have a look at the PassiveDiffusionPl2Int in one of your simulations. You will find that PassiveDiffusionPl2Int is basically never 0 (except for the turning point or when there is no drug in the system any more).
On the other hand, if you set all clearane parameter (hepatic, enzymatic & renal) in your PBPK model to 0 and simulate your system for a long time periode (1000 h or more) you will find that at some point PassiveDiffusionPl2Int will become 0. This is when the system is in equilibrium and the unbound concentration in bothe spaces (here, plasma and interstitial) are equal.
Hope this helps!
Kind regards, Henrik
 Now, I get your idea that unbound concentration between the two spaces (plasma and interstitial) are not equal during equilibrium process, but, another problem comes. I thought Cu + Cb = Ctotal in a compart because V (Cu + Cb)= V Ctotal. However, as the PK-Sim takes account protein as another compart, I think unbound and bound drug don't share the same distribution volume, is it right? And, basing on this change, what is the relationship among Cu, Cb, C_water, C_protein, Vf(water/plasma) and Vf(protein/plasma)?
Now, I get your idea that unbound concentration between the two spaces (plasma and interstitial) are not equal during equilibrium process, but, another problem comes. I thought Cu + Cb = Ctotal in a compart because V (Cu + Cb)= V Ctotal. However, as the PK-Sim takes account protein as another compart, I think unbound and bound drug don't share the same distribution volume, is it right? And, basing on this change, what is the relationship among Cu, Cb, C_water, C_protein, Vf(water/plasma) and Vf(protein/plasma)? and than go to PassiveDiffusionInt2Cell and check the boxes:
and than go to PassiveDiffusionInt2Cell and check the boxes:

 it can deduce that C_plsfu=Cu=C_water=NuV_water_pls, which means fu here is equal to (NuV_pls)/(N_plsV_water_pls). It is not identical to what is generally used that fu=Cu/(Cu+Cb)=Nu/(Nu+Nb)=Nu/N_pls. On the other hand, Cu could be assumed to equal C_water, because plasma almost consists of water.
it can deduce that C_plsfu=Cu=C_water=NuV_water_pls, which means fu here is equal to (NuV_pls)/(N_plsV_water_pls). It is not identical to what is generally used that fu=Cu/(Cu+Cb)=Nu/(Nu+Nb)=Nu/N_pls. On the other hand, Cu could be assumed to equal C_water, because plasma almost consists of water.
Hi, all,
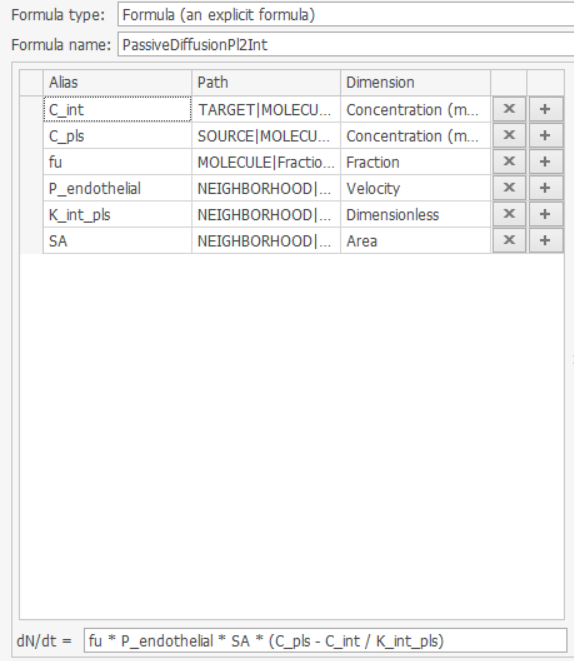
When I learn the formula in MoBi, I find this passive transport PassiveDiffusionPl2Int is defined as "dN/dt=fu P_endothelial SA * (C_pls - C_int / K_int_pls)"; isn't the term "C_pls - C_int / K_int_pls" is equal to 0?