Dear Wilbert,
please find the details how the FcRn mediated recycling for large molecules in PK-sim described here https://link.springer.com/article/10.1007/s10928-017-9559-4
Hope this might help you.
Best, Tobias
Closed wilbertdew closed 4 years ago
Dear Wilbert,
please find the details how the FcRn mediated recycling for large molecules in PK-sim described here https://link.springer.com/article/10.1007/s10928-017-9559-4
Hope this might help you.
Best, Tobias
Dear Wilbert,
yes, you are absolutely right that there is no turnover of FcRn implemented. While the publication that @tobiasK2001 mentions gives you an oversight, it will not help you understand the explicit implementation of what you are trying to do. Section 5 in the Supplementary Information might help you understand how the FcRn SteadyState is defined.
FcRn is located in the "Endogeneous_IgG" Organ. While the initial concentration in the endosome is defined (Parameter "..|Start concentration of free FcRn (endosome)" as a Parameter in the FcRn molecule in you Molecules BuildingBlock), the initial concentrations in Plasma and Interstitium are then calculated based in this value and the initial value of endogenous IgG.
I would thus suggest to create a turnover reaction for FcRn in the endosome of the Endogenous_IgG Organ using "..|Start concentration of free FcRn (endosome)" as the reference steadystate (use "endosome" as "match tag" container criteria.
Let me know if you need further instructions on how to do this.
but I would like to be able to implement it to describe the short half-life of drugs that bind to FcRn at neutral pH as well.
Just out of curiosity: what is the half-life of the drugs you are talking about?
And a happy new year to you, too!
Best, Stephan
Dear Tobias and Stephan,
Thanks a lot for your help! I will try to get this working and will get back with the result :)
The main basis for this shorter half-life with increased binding at pH 7.4 comes from this paper and their figure as inserted, where v3, 4, 5, 6 have increasing affinity at acidic and neutral pH up to 3 and 35 nM, respectively. Any alternative interpretation of these data is also welcome of course :)

That's indeed interesting.
But I am not sure if TMDD through FcRn could be the solution. Is the soluble IL-6R also a FcRn binder (as it seems to have a long half-life by itself, but I could not find anything)? If so: An alternative guess from my side would be that the high-affinity FcRn binders (300-fold increased affinity in comparison to IgG1) would lead to an FcRn saturation which will cause the half-life of all FcRn binders in circulation to drop significantly.
Hi Stephan, sIL-6R should not bind FcRn as in this study sIL-6R concentrations are decreased by the IgGs that bind sIL-6R extracellularly and release it in the endosomes. If it would bind FcRn, this would not be an effective strategy. It might indeed be an option that the increased binding at pH7 causes saturation in the endosomes as well and also shortens half-life of the IgG variant itself as well as its competitors, but at an antibody concentration of around 200 nM, I don't think this is an option given the current estimates of FcRn concentrations.
If it would bind FcRn, this would not be an effective strategy.
If FcRn is saturated, it would be effective. But I agree, that FcRn saturation might be unlikely with the low dose of antibody.
But I also don't see that "normal" turnover of FcRn or Drug-FcRn could be a suitable explanation as other high-affinity FcRn binders (YTE antibodies) show a half-life of 70 days (and more): https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3837853/
However, it might be a compound-specific mechanism that triggerst Drug-Complex-FcRn degradation. Maybe it is the IL-6R.
I haven't checked the publication, but can you exclude binding and TMDD to/at other targets?
The principles of sweeping antibodies are explained here: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5627591/figure/f0001/
These antibodies are designed with pH-dependent binding to both their antigen and FcRn. The antibody collects the antigen outside cells and then the complex binds to FcRn on the cell membrane at pH 7-7.4, initiating cellular uptake and endosomal trafficking. As pH lowers in the endosome, the antigen is released from the antibody but the antibody remains bound to FcRn (ideally). The result is that the antigen is cleared but the antibody is recycled back to the extracellular fluid.
It is worth noting that most antibodies and proteins have negligible binding to FcRn at neutral pH. Their cellular uptake is largely by passive fluid-phase pinocytosis (non-specific). That is why most PBPK models for antibodies only consider FcRn binding in the intracellular endosomes.
The above PK profiles are observed because of increased active uptake into endothelial cells and leukocytes by FcRn binding on the cell membrane at neutral pH without enough recovery in low pH endosomes.
Modeling this with the whole-body PK-Sim framework is near impossible and the uncertainties cloud any potential conclusions. You will need to add pH-dependent antigen binding, pH-dependent FcRn binding, active drug uptake through binding to FcRn on cell membranes and finally endosomal trafficking. I would recommend going with a minimal model + FcRn in MATLAB.
without enough recovery in low pH endosomes.
Yes, the short half-life of e.g. PH-v6 (KD FcRn pH7.4 = 35 nM and KD FcRn pH6 = 3 nM) is what puzzles me. Why would endosomal recovery be reduced?
You raise a great point. I made a poor assumption there. It is certainly plausible that recovery from the endosome is the same for each of these drug scenarios (with identical acidic FcRn affinity). You could even go so far as to approximate elimination with a first order rate constant in the endosome. As the neutral FcRn affinities become stronger, a greater proportion of total drug is sequestered in the endosome and thus the proportion of total drug eliminated over time increases. This would explain the sharper elimination slopes in the plasma PK profile. A sensitivity analysis with a minimal model might be illuminating.
The feature for FcRn binding in plasma doesn't appear to be set up for internalization yet
As a gross alternative, lumping the active (FcRn binding and internalization from the cell membrane) and passive (fluid phase pinocytosis) cellular uptake rates and optimizing the kup parameter while keeping the FcRn affinity at pH 6.0 constant gives you relevant results for the antibody. Modeling the antigen and complex interactions is another story. In this case, kup for PH-v3 is 0.77 1/min and kup for PH-v6 is 2.01 1/min. I used 5.34 nm for hydrodynamic radii. KD at pH 6.0 values extracted from the paper (15 nM and 3 nM respectively).
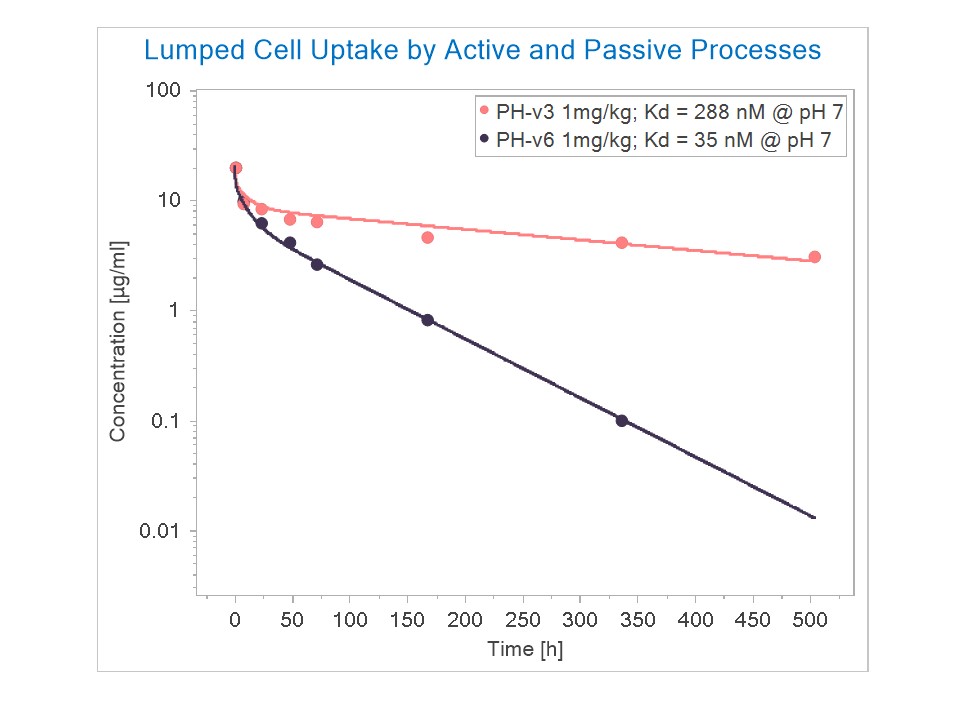
Of course this does not take into account the co-administration of IgG (as in the experiment) but it's a conceptual starting point.
Thanks for these discussions and insights! @StephanSchaller : I agree with you the literature is not so consistent on this topic. The YTE antibodies could have an affinity at pH7 that is still quite low, as in the Yang paper cited by prvmalik, this is determined a 1110 for cyno and 760 nM for human FcRn (but this might differ between IgGs, as the PK effects in their cyno experiments also differ between IgGs). Also, the Robbie paper you referred to states in the intro and discussion that increasing affinity at pH7 and pH6 can decrease half-life, and only increasing affinity at pH6 is required. The half-life of FcRn itself is believed to be around 10h only, probably caused by a small fraction that ends up in the lysosome and the fast circulation of FcRn (sorry, don't have the literature available for FcRn half-life). I agree that the fraction of endosomal recovery doesn't need to be reduced. @prvmalik : Thanks for your considerations on the possibility to model this behaviour in PK-Sim. I agree that the whole sweeping of target by these kind of antibodies might be to difficult. I would like to focus mainly on the PK of the antibody itself. (Also, I am wondering if missing this FcRn degradation in the model might give a misprediction of biodistribution data for normal antibodies with residualizing labels, but this is not the main focus) Your simulation approach is rather interesting for me, I had not considered that option. The interstitial Kd in your screenshot is in the uM unit, can you get similar results with 288 nM or did you already change it before simulating? I would still like to try to incorporate degradation of FcRn to see how far I can get and to enable prediction from in vitro affinity data. Should it be impossible to get increased internalization with interstitial FcRn binding in current PK-Sim model after inclusion of FcRn degradation? Currently with implementing FcRn degradation in the reactions building block, I do get a sensitivity to interstitial FcRn Kd towards half-life, but I did not use container criteria yet, and using the "..|Start concentration of free FcRn (endosome)" for synthesis effectively reduces the total concentration to the initial free concentration, I am afraid.
Yes Wilbert, I did use 288 nM for the test but the feature does not work as intended. I created the screenshot afterward (thanks it is fixed now!). I haven't yet opened up Mobi to look at what the feature is doing but (based on the resulting PK profile) I imagine it is just binding and sequestering at the cell membrane rather than internalizing and trafficking to the intracellular endosomes.
The profiles I simulated are WITHOUT using this feature (i.e. KD FcRn in plasma/int is 999999 uM as default), only lumping the active and passive cellular uptake into the kup parameter.
FcRn degradation and turnover on the cell membrane would only be relevant to include in the model if binding of the antibody induced internalization. However what is more likely (as in the case of HER2 for instance) is that the mAb-FcRn receptor complex gets internalized according to the natural turnover rate of FcRn. Many membrane-bound receptors are in a constant flux from the membrane and back to the endosomes/cytoplasm as the cell membrane is quite dynamic in nature. Since every time an old FcRn receptor (or complex) is internalized it is simultaneously replaced with a new naked FcRn receptor, you can model kon/koff binding and a first order internalization rate for the complex without having to model the turnover explicitly (see the approximation example in Shah 2012 and in my 2017 trastuzumab paper).
From what I understand of what you have done, interstitial/cell membrane FcRn binding and subsequent degradation of the complex is another way of approximating the situation however then you cannot use the in vitro values for the pH 6 binding - the intracellular processes end up being lumped into the first order degradation of the complex. This would throw a reviewer for a loop because you still have passive uptake and intracellular FcRn binding going on that is not linked.
Here's what we're dealing with:
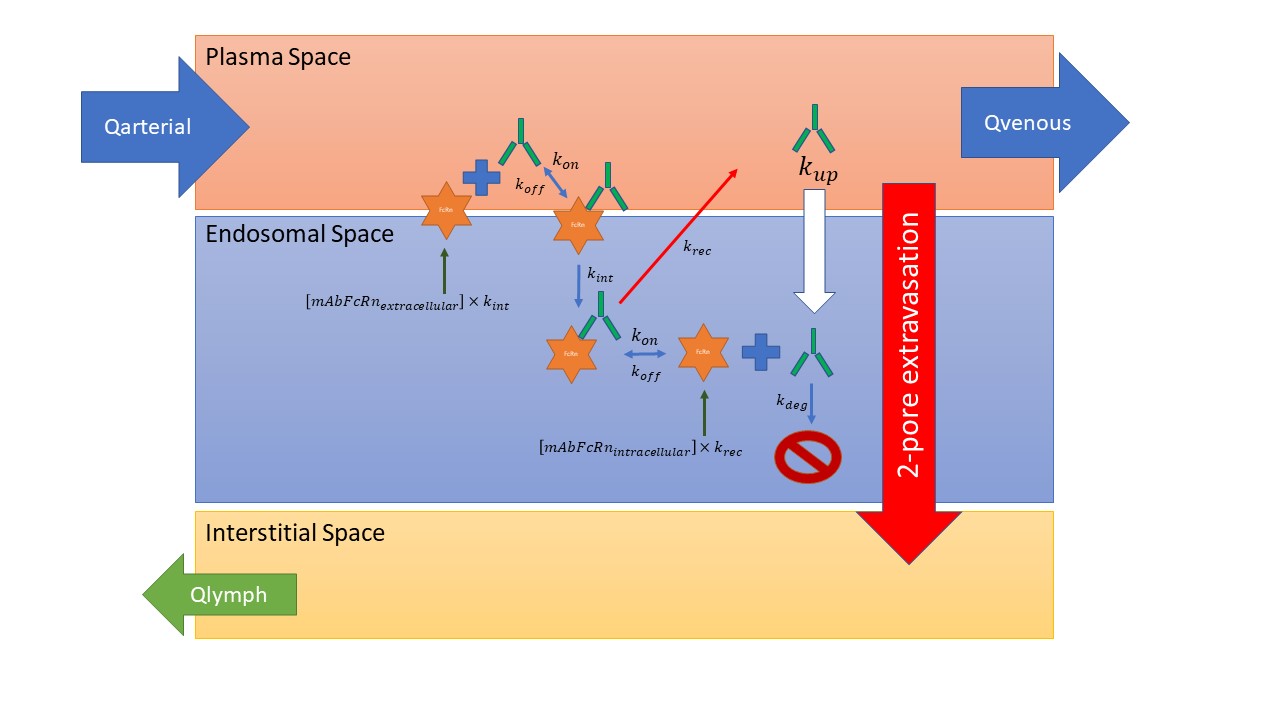
Mobi is challenging for this type of system because it only accepts passive transport of molecular species for rates; in other words to maintain a constant FcRn on the surface you will be replenishing it with [extracellular mAb-FcRn complex] x kint.
I haven't yet opened up Mobi to look at what the feature is doing but (based on the resulting PK profile)
@prvmalik How did you change kup in PK-Sim if you haven't opened MoBi, yet? This parameter should only be accessible in MoBi, no?
As a gross alternative, lumping the active (FcRn binding and internalization from the cell membrane) and passive (fluid phase pinocytosis) cellular uptake rates and optimizing the kup parameter while keeping the FcRn affinity at pH 6.0 constant gives you relevant results for the antibody.
The profiles I simulated are WITHOUT using this feature (i.e. KD FcRn in plasma/int is 999999 uM as default), only lumping the active and passive cellular uptake into the kup parameter.
@prvmalik can you elaborate a bit on how you are lumping these processes? Per default PK-Sim does not consider internalization of the drug-FcRn complex:

Are you thus reducing the recycling rate (NetMassTransfer_EndosomalToInt/Pls_FcRn_Complex) if you want to consider an increased drug-FcRn Internalization?
Also, in our discussion here, we might confuse internalization of FcRn and recycling vs FcRn degradation (actual molecular degradation and re-synthesis).
Thanks for your questions Dr. Schaller.
You can access the kup parameter in PK-Sim by viewing "advanced" parameters under the "vascular physiology" tab.
As for the lumping, process 1 (kon, koff, subsequent kint) is lumped with process 2 (kup) translating to net uptake of unbound mAb into the endosomal space. Of course, as I said before this is a gross approximation because what you end up with is only unbound mAb being taken up rather than unbound mAb (from kup) and FcRn-bound mAb (from kint). However if affinity is sufficiently strong it may be possible to assume that equilibrium is quickly reached at the new pH 6 affinity in the endosome.
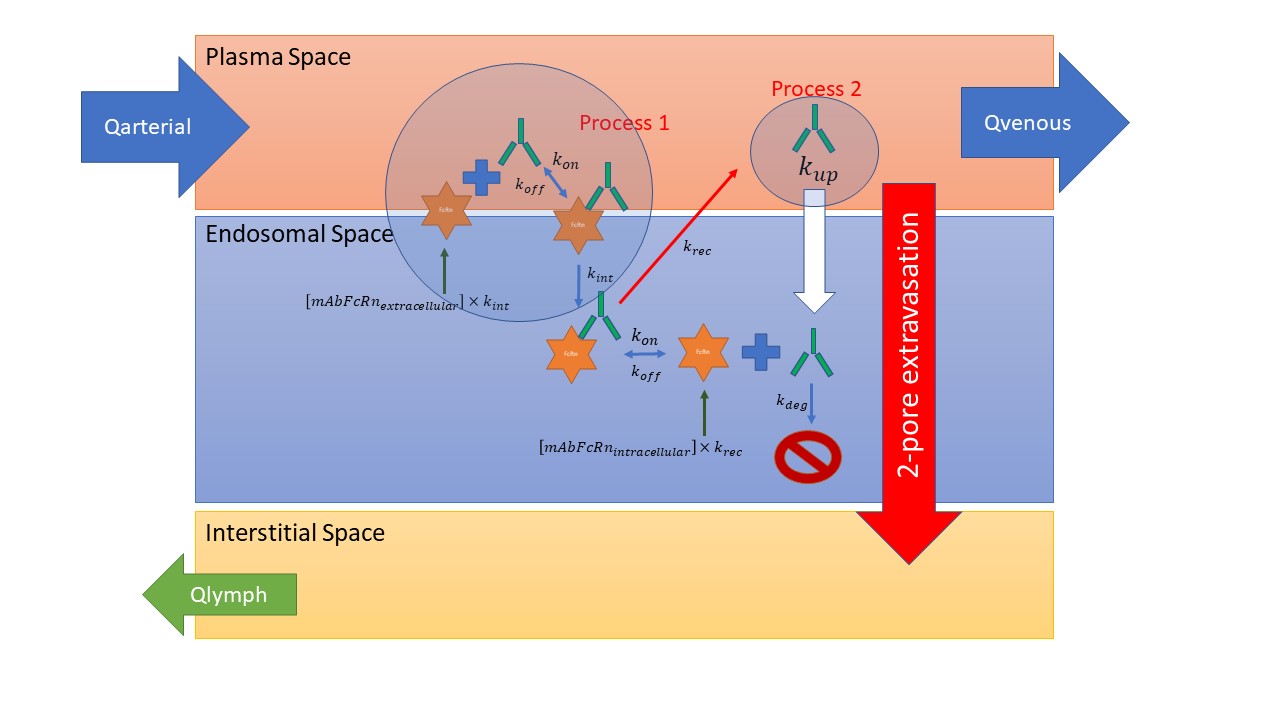
The normal FcRn steady state kinetics at pH 6 inside the endosome remain unchanged. I only increased the kup parameter numerically to reflect that more drug is entering the endosome by binding to FcRn on the cell membrane and subsequent internalization
You can access the kup parameter in PK-Sim by viewing "advanced" parameters under the "vascular physiology" tab.
Thanks... must have missed this one.
As for MoBi, Drug-FcRn-Complex can be transported by passive transports (as it is not a "stationary" molecule).
Of course, as I said before this is a gross approximation because what you end up with is only unbound mAb being taken up rather than unbound mAb (from kup) and FcRn-bound mAb (from kint).
Yes, an approximation but a quick shortcut.
In MoBi you could substract the [extracellular mAb-FcRn complex] x kint term from the existing formula in the NetMassTransfer_EndosomalToInt/Pls_FcRn_Complex trannsports (be sure to use "TARGET|MOLECULE|Concentration" and just be aware you might need to reduce K_uptake ("..|Rate constant for endosomal uptake") accordingly).
@prvmalik you might have mixed up [extracellular mAb-FcRn complex] x kint and [extracellular mAb-FcRn complex] x krec in your drawing above.
Thanks @prvmalik for including these clarifying figures. There are a few things I still don't get. The current implementation in PK-Sim as I understand it looks in a schematic way as depicted below

Compared to the scheme from the publication as included by Stephan, there are a few extra processes in my scheme:
Are these processes not included in the PK-Sim model or just not in the published scheme? If they are not included in the model, is this just a simplification of the equations while keeping the same behaviour or are there some consequences for molecules binding at pH7?
You are saying "Mobi is challenging for this type of system because it only accepts passive transport of molecular species for rates; in other words to maintain a constant FcRn on the surface you will be replenishing it with [extracellular mAb-FcRn complex] x kint." I do not understand this completely, in the reactions building block, you can implement any order of reactions I thought, can we not include synthesis as a reaction (rather than transport) in a way as depicted below?

The initial endosomal FcRn concentration should be equal to the total endosomal FcRn concentration as kdeg, kint and krec are equal for free and mab-bound FcRn. If this concentration is not available as parameter, we can just extract the value from a simulation without degradation in Mobi. It would make more sense physiologically to implement the synthesis to the membrane bound FcRn, maybe, but I don't think this will alter the behaviour of the model.
Sorry for the confusion on my side, please let me know if you think this discussion is running in circles and not progressing anymore :)
Are these processes not included in the PK-Sim model or just not in the published scheme? If they are not included in the model, is this just a simplification of the equations while keeping the same behaviour or are there some consequences for molecules binding at pH7?
Recycling of the free FcRn is included and indeed not correctly depicted in the figure i posted. However, internalization of the Drug-FcRn complex is not. It seems this was omitted as the FcRn module was not forseen to be used for pH7-FcRn binders (they were also not part of the validation data set).
Assuming a significant fraction of FcRn on the membrane is bound to drug and the internalization rate is the same as for the unbound FcRn this could have consequences for molecules binding at pH7.
Thanks @StephanSchaller that helps a lot! And we probably cannot implement it ourselves as only passive transport is possible, as indicated by @prvmalik
@wilbertdew , I am not sure if maybe @prvmalik meant sth. else but within MoBi this could be implemented by simply substracting the missing term from the exocytosis transport reaction for the Drug-FcRn complex as I described above:
In MoBi you could substract the [extracellular mAb-FcRn complex] x kint term from the existing formula in the NetMassTransfer_EndosomalToInt/Pls_FcRn_Complex trannsports (be sure to use "TARGET|MOLECULE|Concentration" and just be aware you might need to reduce K_uptake ("..|Rate constant for endosomal uptake") accordingly).
Yes, I trust that it will be able to be done as Stephan has described. What I meant was that it's more complicated in Mobi because you have to use the 'Passive Transport' building blocks (this is their name, not necessarily meaning passive transport literally).
@prvmalik you might have mixed up [extracellular mAb-FcRn complex] x kint and [extracellular mAb-FcRn complex] x krec in your drawing above.
Yes it will take some finessing in Mobi to get mass balance correct
@wilbertdew , how did you solve your issue in the end? Thanks, Stephan
Hi @StephanSchaller,
I am still working on it when I have time, but it seems I can include bidirectional transport for each molecule by extending the selection of molecules of each monodirectional transport that is already present in the model. It is a bit of work to recalculate the parameter values from the original model and I am documenting all steps so hopefully I can share my solution in more detail soon. Best,
Wilbert
Hi @wilbertdew , thanks. Good luck and looking forward to hearing how your solution will work.
Best, Stephan
Hey everyone,
The discussion on this forum helped me a lot to properly look into it and work on a solution for the identified problem. The results are now documented and published, you can find a view-only link with model files here. We will make the model files available on GitHub as well. Feel free to comment, question or discuss this further. @msevestre Maybe this can be moved to a discussion also to make it more easy to find back?
Hi All,
Can anybody give me some pointers to better underdstand FcRn kinetics in PK-Sim/Mobi? Currently, I have the impression that no turnover of FcRn or degradation for the drug-FcRn complex is implemented. I do understand that FcRn binding is meant to minimize this degradation, but I would like to be able to implement it to describe the short half-life of drugs that bind to FcRn at neutral pH as well. In Mobi, I can implement degradation of the complex, but then I also want to implement degradation of FcRn itself (to avoid changing FcRn concentrations as a consequence of drug binding) for which I need to compensate with FcRn synthesis. I am now in doubt whether I need to use the endosomal or the membrane concentration of FcRn to calculate synthesis rate and if the latter is actually a parameter in the model. Also, I cannot find the reactions in mobi describing transfer of FcRn to endosome, or is this not defined as a reaction? Thanks for your help and already a happy new year to everyone! Wilbert