As I understand it, using a tablet for example, after ingestion the dissolution of the drug happens according to particle size groups and Noyes-Whitney.
It's true if the formulation chosen was Particle Dissolution. For other formulation types a corresponding dissolution function is used (Weibull, Lint80, ...)
Is this transit function referring to the Gaussian distribution model? I presume not because, as I understand it, the compartmental model replaced this one
Correct
I've tried to understand the model structure by exporting PK Sim models to .txt and also by using MoBi to export to Matlab .m files but I haven't yet figured it out. I'm particular interested in the general form of these processes (i.e. given an SI compartment i, the transfer and absorption in that compartment), so if someone could point me in the direction of the model equations, I would be very appreciative!
You can find the formulas of the drug transfer along the lumen and (passive) drug absorption from the lumen into mucosa in the Passive Transports building block in MoBi. Both applies to the liquid drug (=dissolved part of a tablet)
LumenDrugTransport_xxx describes the drug transfer along the lumen.
E.g. LumenDrugTransport_duo describes drug transfer Lumen/Stomach :arrow_right: Lumen/Duodenum
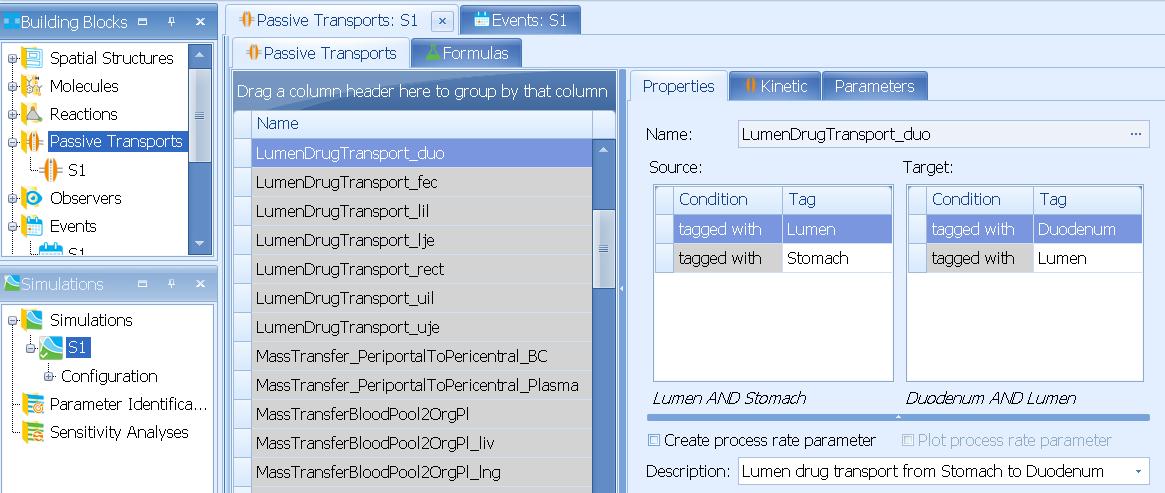
Under Kinetic you can see the corresponding process rate:

DrugAbsorption_trans and DrugAbsorption_para processes describe (passive) transcellular and paracellular absorption from Lumen into Intestinal Mucosa. In the current model paracellular absorption is always zero per default.
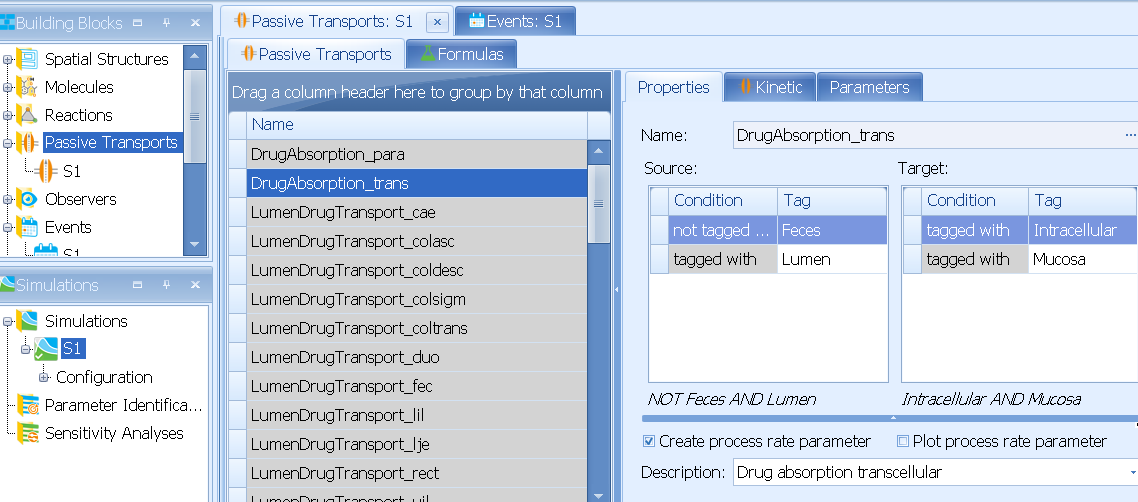
If you look at the kinetic of DrugAbsorption_trans you will see that it points to the NEIGHBORHOOD|MOLECULE|Drug absorption rate (lumen to mucosa) parameter.

Here, the NEIGHBORHOOD keyword refers to the neighborhood between source and target compartment of the transport, and you will find the final equation in the corresponding neighborhoods of the spatial structure building block, e.g Neighborhood between Lumen/Duodenum and Mucosa/Duodenum/Intracellular(please refer to the user manual for more details about using keywords in formulas):

If some active transport processes are available in your model, you can find process description e.g. in the Molecules building block:

Movement of the tablet along the lumen and release rate of the tablet is described in the Events building block. Release rate depends on the formulation type.
E.g. for the formulation type Particle Dissolution: you have transports ParticlesDrugReleaseXXX
Each transport describes the dissolution process of particles in one lumen segment and is defined as
Release rate fraction XXX * Release rate of the tablet

Release rate of the tablet depends on the formulation type. E.g. for particles dissolution it's described by the Noyes-Whitney equation.

Release rate fraction XXX is the result of the movement of the tablet/particles along the lumen (Stomach :arrow_right: Duodenum :arrow_right: ...) and reflects the fraction of tablet/particles in the corresponding lumen segment.

s. example project attached Example_OralApplication_MonoParticles.zip
Sorry in advance if this is a very basic question. I'm trying to discern the underlying model equations of transit between the lumen and the mucosa, and the absorption from the mucosa to the portal vein used in PK Sim. I have read both Thelen papers (Thelen et al. 2011, J. Pharm Sci; Thelen et al. 2012, J. Pharm. Sci) and the third part by Willmann (Willmann, Thelen, and Lippert 2012, J. Pharm and Pharmacol), but it remains unclear to me what specific equations regulate these processes.
As I understand it, using a tablet for example, after ingestion the dissolution of the drug happens according to particle size groups and Noyes-Whitney. Then, from Thelen (2011), "the drug is transported along the lumen of the intestine according to the transit function and can enter the mucosa of the corresponding segment either by the transcellular route directly into the enterocytes or by paracellular diffusion into the intersitial space of the mucosa...The fully dissolved drug once absorbed into the mucosal space can then diffuse from one sub-compartment to another, can be metabolized in the intracellular space, and finally reaches the vascular space of the mucosa to enter the portal vein blood."
Is this transit function referring to the Gaussian distribution model? I presume not because, as I understand it, the compartmental model replaced this one to alleviate technical complications. I have experience with other PBPK intestinal absorption models (e.g. Fenneteau, Poulin, and Nekka 2010, J. Pharm Sci; Peters 2008, Clin Pharmacokinet; Agoram, Woltosz, Bolger 2011, Adv. Drug Deliv. Rev.; Gertz, Houston, and Galetin 2011, Drug Met Disp; Yu and Amidon 1999, Int J Pharm etc.) so I have some handle on these processes are modelled therein, does PK Sim's compartment model compare with them (modulo the inclusion of internal compartments in each organ, like the RBCs etc., of course)?
I've tried to understand the model structure by exporting PK Sim models to .txt and also by using MoBi to export to Matlab .m files but I haven't yet figured it out. I'm particular interested in the general form of these processes (i.e. given an SI compartment i, the transfer and absorption in that compartment), so if someone could point me in the direction of the model equations, I would be very appreciative!