Hi @teutonicod,
The plasma clearance is as far as I can tell from your description the input parameter from the clearance process. e.g. this is what the user enters in the system. PK-Sim then calculates, using a well-stirred model, the specific clearance. see here for details
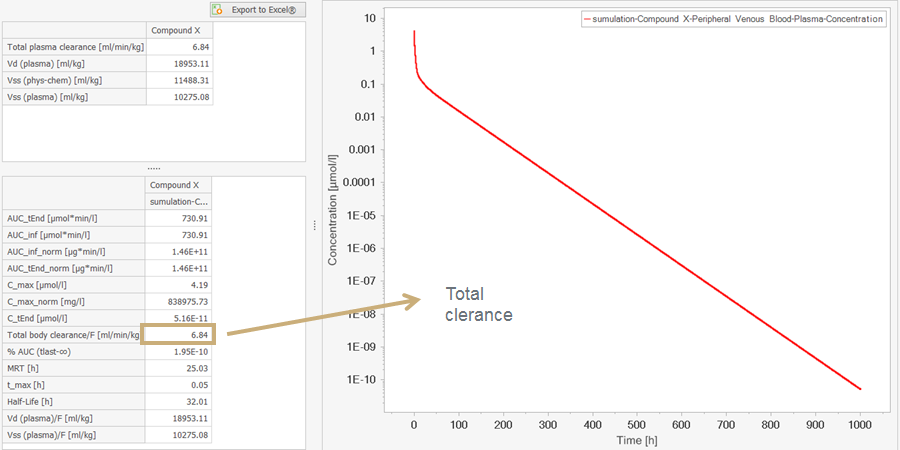
In order to calculate the Plasma clearance in the PK-Analysis section, PK-Sim uses the last 10% of the curve. Please make sure you have simulated long enough. My suggestion would be to increase your simulation time and then check to see if the values are similar
Hope that helps









Dear all,
In the unspecified liver clearance process in PK-Sim, the intrinsic clearance for the liver is calculated using several parameters, one of them being referred to as Plasma clearance. Do you know how this parameter is defined? In a simple model (IV bolus and only unspecified clearance) the value for this Plasma clearance does not coincide with the Total body clearance in the PK-Analysis section, do you have an explanation for the difference of these parameters? Thanks you in advance for your help!
Best regards,
Donato