I suspect these installation errors may be coming from the default setting of R regarding saving and loading of workspaces. This would cause loaded packages to stay loaded even if you start RStudio in a supposedly clean session.
I strongly recommend go to the Global Options of RStudio and de-select "load .RData on startup" and set "Save workspace to .RData on exit" to "Never" as illustrated in this forum post from Hadley:

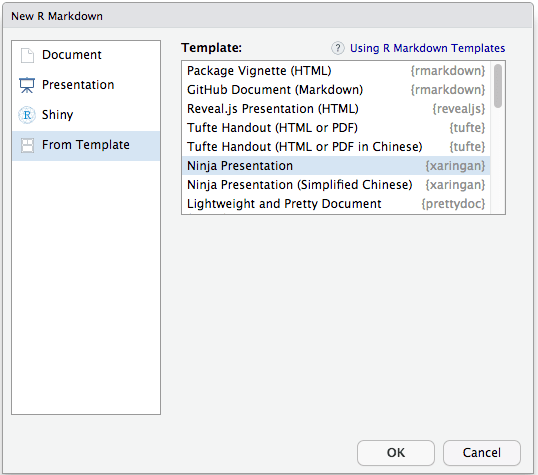
Walking through installing from github and he got a whole bunch of errors - starting with not being able to update packages. See below.
Probs good to address next week before sending out to epis.
He needed to reinstall the package colorspace and restart the R session and then it worked for colorspace. Then got same error for rlang, repeated above steps and then epireports loaded correctly but templates are still not there.... @dirkschumacher @zkamvar