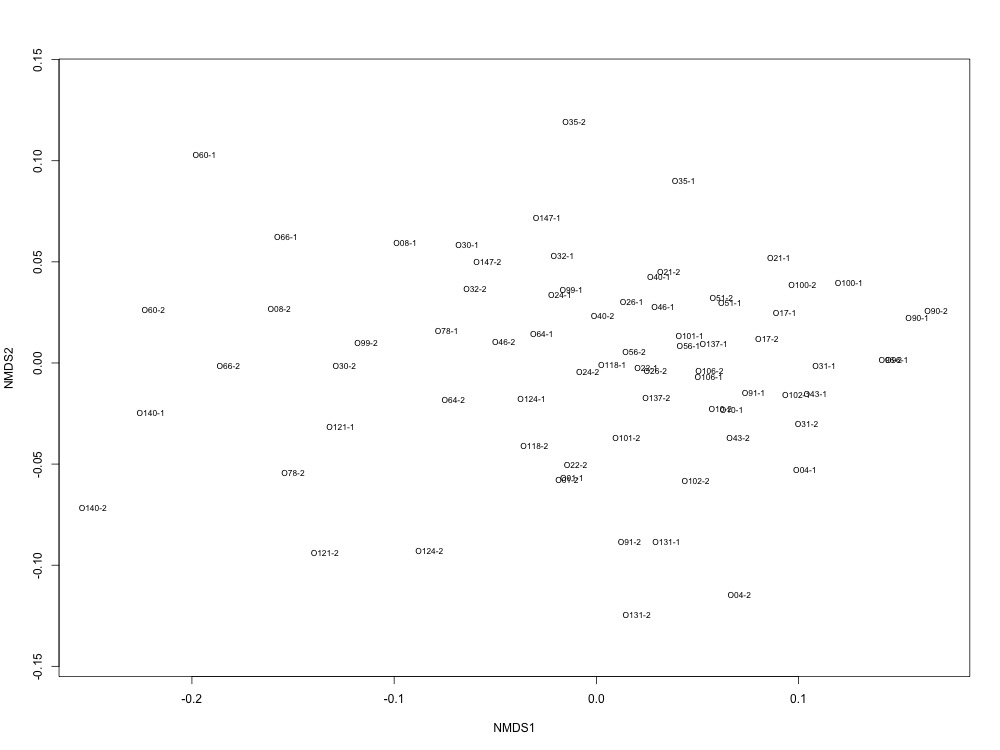
I need a bit of clarification: Are you saying that samples with poor technical replication had both replicates run on the first column? Or had one replicate run on the first and one of the second column? Either way, if you are confident that the source of poor replication is the column, then there isn't a reason to rerun your samples. However, if I remember correctly, none of your samples showed the kind of technical replication that we expected so we are actually look at different levels of bad replication. Is that correct? If replicates run on the second column were also less than ideal, then there is still reason to rerun a couple and see if we can do better.
Notebook
Looks like the samples with poor technical replication were run on the first column. The second technical replicate of those samples were the sample we ran before we changed the column. My guess is the technical issue with the column affected those samples before we realized we needed to change the column.
@emmats I can get you 2 of those samples today or tomorrow! When's a good time to drop by Genome Sciences?