@mgavery here is a draft (version 1.1)
https://github.com/sr320/nb-2021/blob/main/C_gigas/analyses/top.annotv1.1.tab
Closed sr320 closed 3 years ago
@mgavery here is a draft (version 1.1)
https://github.com/sr320/nb-2021/blob/main/C_gigas/analyses/top.annotv1.1.tab
here is a(version 1.2 ) - with sea urchin gene symbols
https://github.com/sr320/nb-2021/blob/main/C_gigas/analyses/top.annotv1.2.tab
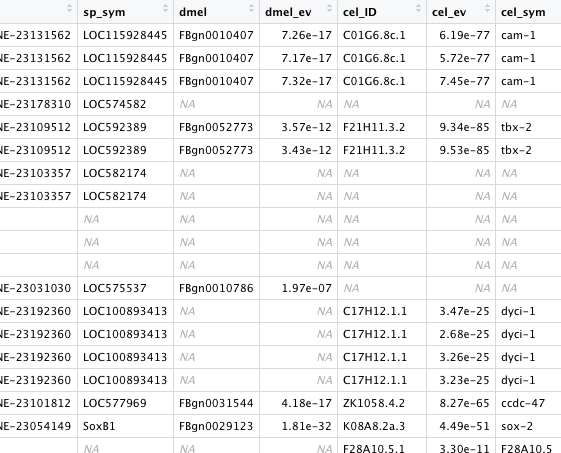
There are now tables at https://github.com/sr320/nb-2021/tree/main/C_gigas/analyses
Where select supplemental tables from Spur scRNA-seq paper are joined with top markers..
specifically
here is a(version 1.3 ) - with sea urchin gene symbols + no evalue limit on sea urchin blast.
https://github.com/sr320/nb-2021/blob/main/C_gigas/analyses/top.annotv1.3.tab
There are now tables at https://github.com/sr320/nb-2021/tree/main/C_gigas/analyses
Where select supplemental tables from Spur scRNA-seq paper are joined with top markers.. using 1.3 annotations.
Based on SC RNA-seq here is a gene table to annotate based on genes in model species. etc. topmarker_bla_mitov7_top10perCluster.txt