With sea star data, ran 1) Hisat with genome, 2) Kallisto with cds as defined by genome, and 3) Kallisto with 10 year old transcriptome.
Exact same reads.
Alignments
1) Hisat with genome
2) Kallisto with cds as defined by genome
3) Kallisto with 10 year old transcriptome
There is a lot going on here -
a) why do some samples have reduced alignment only when associated with new genome? (1 and 2)
b) why is there such a big difference in 1) and 2) with respect to alignment rate?
c) what is next step?
With sea star data, ran 1) Hisat with genome, 2) Kallisto with cds as defined by genome, and 3) Kallisto with 10 year old transcriptome. Exact same reads.
Alignments 1) Hisat with genome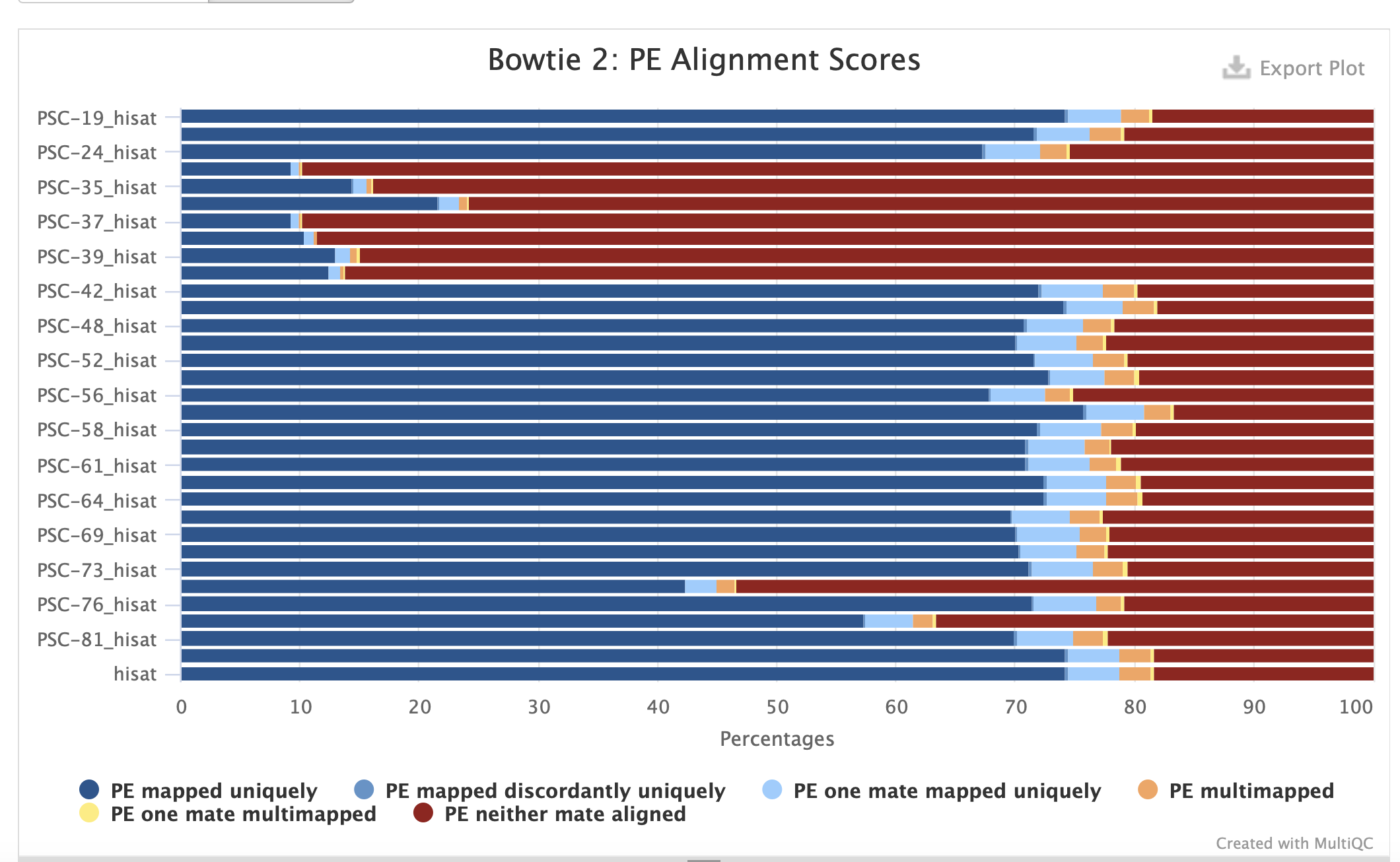
2) Kallisto with cds as defined by genome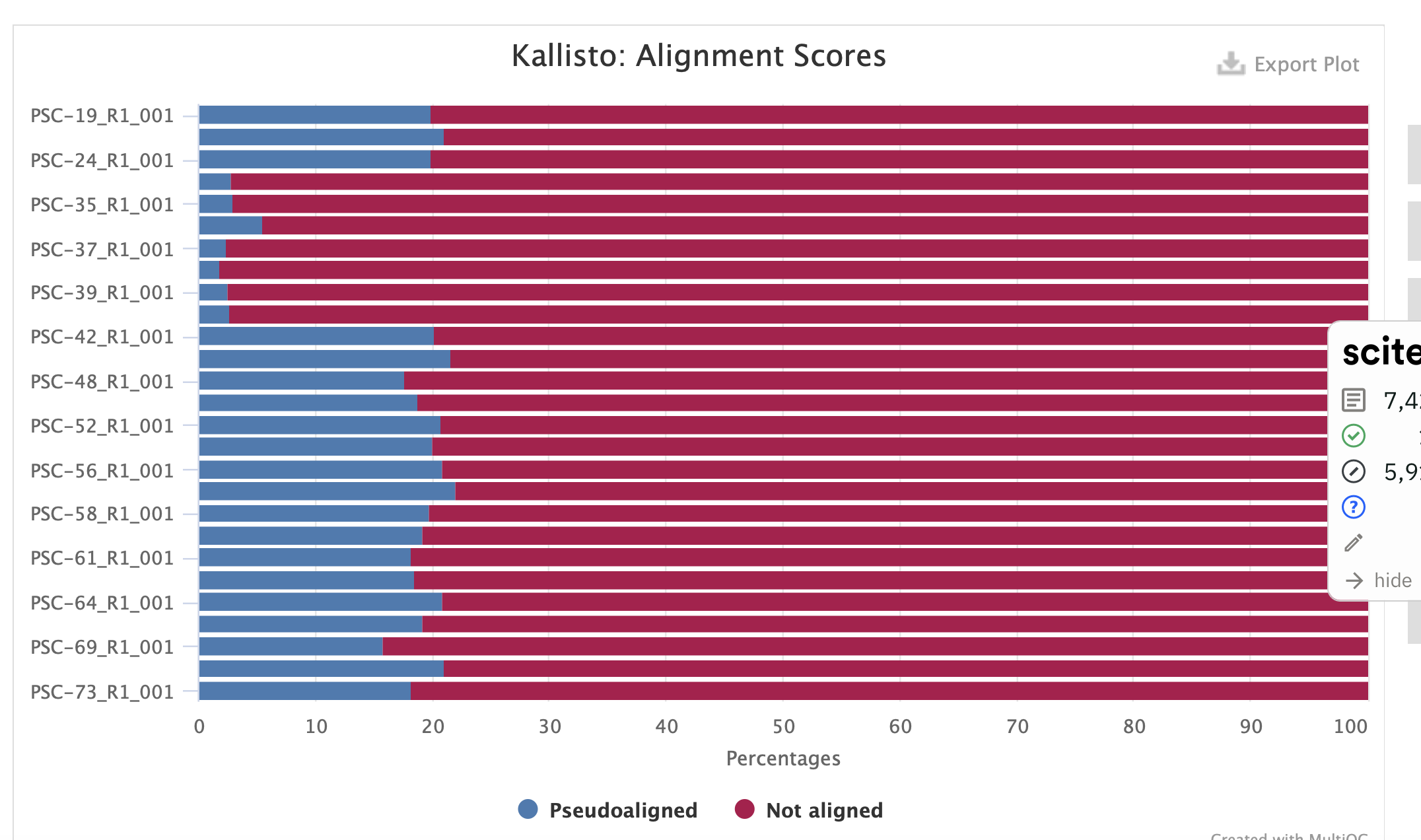
3) Kallisto with 10 year old transcriptome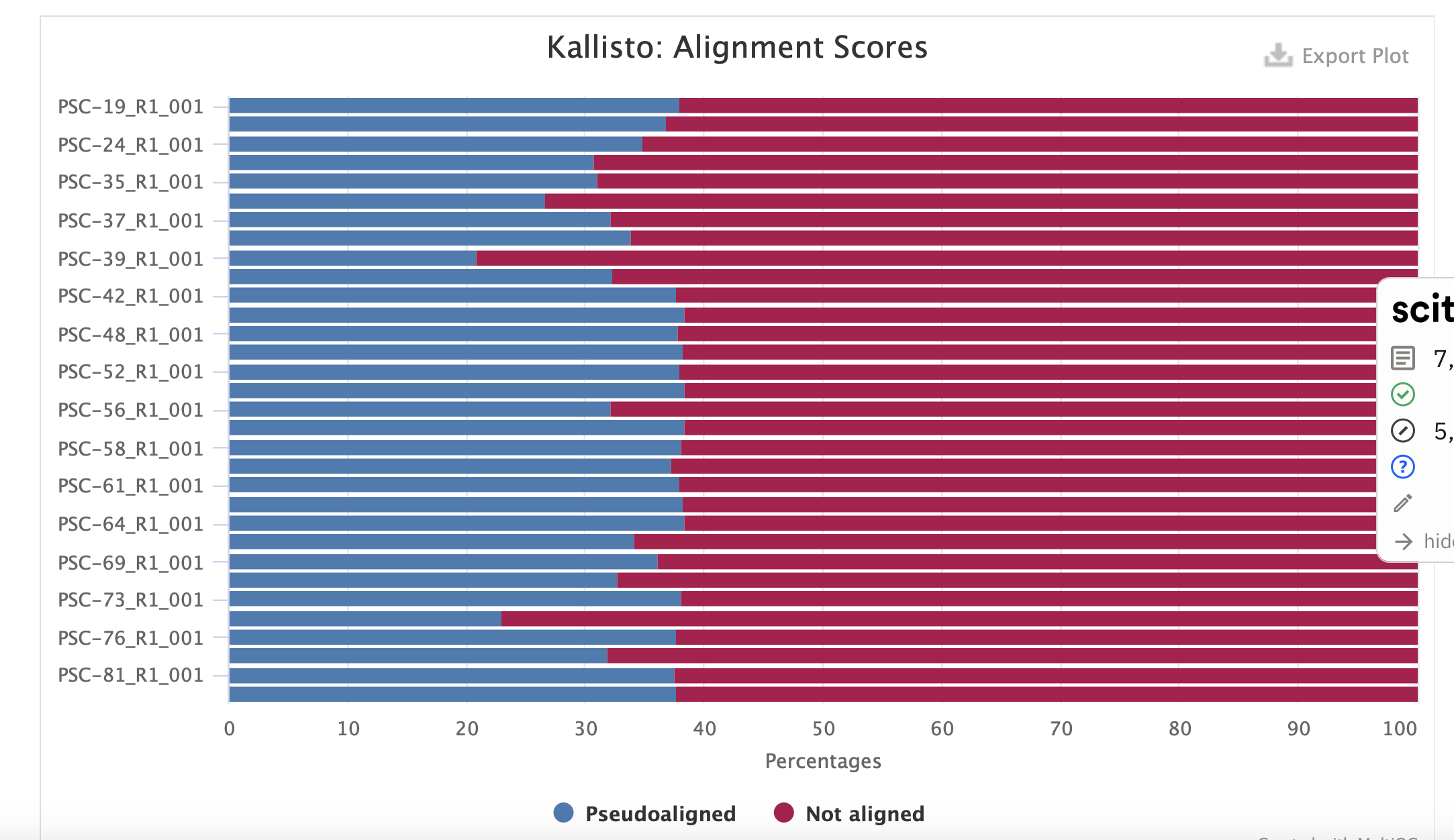
There is a lot going on here -
a) why do some samples have reduced alignment only when associated with new genome? (1 and 2) b) why is there such a big difference in 1) and 2) with respect to alignment rate? c) what is next step?