For the 12 libraries indicate the quantity or RNA added to the 12 pools.
On Thu, Jun 7, 2018 at 3:12 PM grace-ac notifications@github.com wrote:
Cool beans! How?
Related: this is my notebook post https://github.com/grace-ac/grace-ac.github.io/blob/master/_posts/2018-06-03-samples-for-seq.md on my selection for pooling... Has explanation of pooling schema shown below.
Pooling schema image (pools are different colors (n=12)): [image: img] https://camo.githubusercontent.com/c7748919496ad3081f694ac54edd9d097c72f0af/687474703a2f2f6f776c2e666973682e77617368696e67746f6e2e6564752f736361706861706f64612f67726163652f437261622d70726f6a6563742f32303138303630372d73616d706c65732d666f722d7365712e706e67
Note 1: The Day 9 infected (dark orange) and uninfected (light orange) pools were picked such that there are 4 crabs/pool (because the other pools are 4 crabs) and based on the Avg sq day 01 value and I picked the lowest for the uninfected and the highest for the infected (orange boxes corresponding to day 9 pools)
Note 2: I can try and do this in R if that is preferred.
— You are receiving this because you authored the thread. Reply to this email directly, view it on GitHub https://github.com/RobertsLab/resources/issues/285#issuecomment-395581967, or mute the thread https://github.com/notifications/unsubscribe-auth/AEPHt54Ar32dD9wLP2HRQfDKDDFrsV_Cks5t6aUbgaJpZM4UfKt_ .








Cool beans! How?
Related: this is my notebook post on my selection for pooling... Has explanation of pooling schema shown below.
Pooling schema image (pools are different colors (n=12)):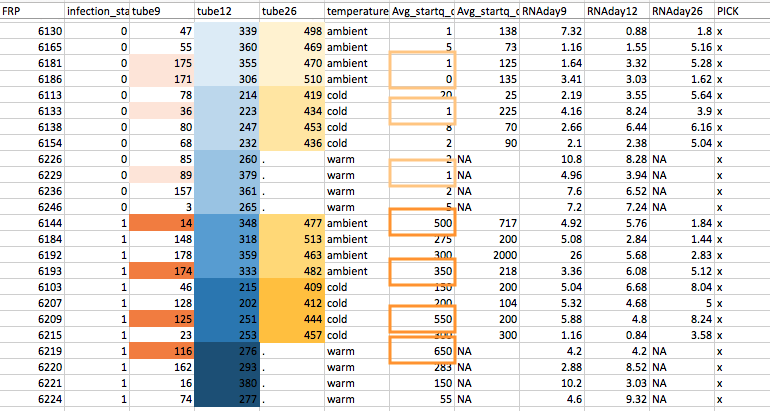
Note 1: The Day 9 infected (dark orange) and uninfected (light orange) pools were picked such that there are 4 crabs/pool (because the other pools are 4 crabs) and based on the Avg sq day 01 value and I picked the lowest for the uninfected and the highest for the infected (orange boxes corresponding to day 9 pools)
Note 2: I can try and do this in R if that is preferred.