Here is file with GO information https://d.pr/f/TVZROq
Closed sr320 closed 5 years ago
Here is file with GO information https://d.pr/f/TVZROq
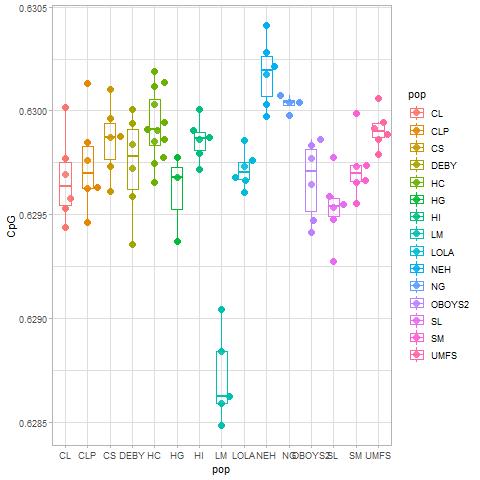
For the plots below:
Note that both graphs below will have the same number of points because the points are the samples/populations, and there are always 90 samples. The number of values represented underneath the points, e.g., the genes, is what changes between the "all samples" and "stress response" plots.
 All genes= left column
Stress genes= right column
All genes= left column
Stress genes= right column
 All genes plot.
All genes plot.
 Stress response plot. Only genes that merged to stress response in the GO information file are represented here.
Stress response plot. Only genes that merged to stress response in the GO information file are represented here.
I can easily put genes on the x-axis instead of samples. I just wasn't sure what would be more valuable. If the samples represent the same treatment, population, or replicates then I think genes on the x axis makes more sense. However, if the question is what differences are occurring between samples/populations/etc. than samples on the x-axis would probably make more sense.
Samples/populations should be on the x axis because there are over 39k genes (or in the case of stress response ~3k) and that would be illegible.
Edit: Misunderstood what values were represented in the original dataset- corrected explanations.
Can you give me a list of stress genes where I could pick one and you could plot the CpG O/E for that for all 91 samples?
I just want to clarify- are rows or columns samples?
columns - 90
thanks Steven On Mar 20, 2019, 2:45 PM -0700, Kaitlyn Mitchell notifications@github.com, wrote:
I just want to clarify- are rows or columns samples? — You are receiving this because you authored the thread. Reply to this email directly, view it on GitHub, or mute the thread.
So the rows are genes? When making the graphs I assumed that the columns were genes and rows were sample because the columns were names of genes, but I can update the description.
Yes
thanks Steven
Steven B. Roberts Kenneth K. Chew Endowed Professor School of Aquatic and Fishery Sciences University of Washington, Seattle WA sr320@uw.edu - 206.866.5141 robertslab.info - @sr320 On Mar 20, 2019, 3:56 PM -0700, Kaitlyn Mitchell notifications@github.com, wrote:
So the rows are genes? When making the graphs I assumed that the columns were genes and rows were sample because the columns were names of genes, but I can update the description. — You are receiving this because you authored the thread. Reply to this email directly, view it on GitHub, or mute the thread.
How about a swarm plot of CpG OE of all stress response genes for all samples
This is on stress genes.
I used three graphs to distinguish differences between the population: density, box and swarm plots





Could you also plot on with these GO ID
GO:0100044 negative regulation of cellular hyperosmotic salinity response by transcription from RNA polymerase II promoter
GO:0042538 hyperosmotic salinity response
GO:0042539 hypotonic salinity response
GO:1900464 negative regulation of cellular hyperosmotic salinity response by negative regulation of transcription from RNA polymerase II promoter
GO:1900069 regulation of cellular hyperosmotic salinity response
GO:1900070 negative regulation of cellular hyperosmotic salinity response
GO:0071477 cellular hypotonic salinity response
GO:0071475 cellular hyperosmotic salinity response
GO:1902074 response to salt
GO:1902075 cellular response to salt
GO:0009651 response to salt stress
GO:1901000 regulation of response to salt stress
GO:1901002 positive regulation of response to salt stress
GO:1901001 negative regulation of response to salt stress
GO:1901200 negative regulation of calcium ion transport into cytosol involved in cellular response to salt stress
GO:1901196 positive regulation of calcium-mediated signaling involved in cellular response to salt stress
GO:1901199 positive regulation of calcium ion transport into cytosol involved in cellular response to salt stress
GO:0071472 cellular response to salt stressAnd separately these GO IDs
GO:0070415 trehalose metabolism in response to cold stress
GO:0070414 trehalose metabolism in response to heat stress
GO:0090441 trehalose biosynthesis in response to heat stress
GO:0090442 trehalose catabolism in response to heat stress
GO:0061408 positive regulation of transcription from RNA polymerase II promoter in response to heat stress
GO:1990845 adaptive thermogenesis
GO:0031990 mRNA export from nucleus in response to heat stress
GO:0043620 regulation of DNA-templated transcription in response to stress
GO:0043555 regulation of translation in response to stress
GO:1990611 regulation of cytoplasmic translational initiation in response to stress
GO:1990497 regulation of cytoplasmic translation in response to stress
GO:0034605 cellular response to heat
GO:0070417 cellular response to cold
GO:0006950 response to stress
GO:0036003 positive regulation of transcription from RNA polymerase II promoter in response to stress
GO:0043618 regulation of transcription from RNA polymerase II promoter in response to stress
GO:0080134 regulation of response to stress
GO:0080135 regulation of cellular response to stress
GO:0097201 negative regulation of transcription from RNA polymerase II promoter in response to stress
GO:0033554 cellular response to stress
GO:0033555 multicellular organismal response to stress
GO:0009271 phage shockand
GO:0045087 innate immune response
GO:0061760 antifungal innate immune response
GO:0045088 regulation of innate immune response
GO:0002218 activation of innate immune response
GO:0002227 innate immune response in mucosa
GO:1905036 positive regulation of antifungal innate immune response
GO:1905035 negative regulation of antifungal innate immune response
GO:1905034 regulation of antifungal innate immune response
GO:0035420 MAPK cascade involved in innate immune response
GO:0045824 negative regulation of innate immune response
GO:0045089 positive regulation of innate immune response
GO:0002758 innate immune response-activating signal transduction
GO:0002766 innate immune response-inhibiting signal transduction
GO:0090714 innate immunity memory response
GO:0035419 activation of MAPK activity involved in innate immune response
GO:0035423 inactivation of MAPK activity involved in innate immune response
GO:0035422 activation of MAPKKK activity involved in innate immune response
GO:0035421 activation of MAPKK activity involved in innate immune response
GO:0046735 passive induction of host innate immune response by virus
GO:0046738 active induction of innate immune response in host by virus
GO:0038075 MAP kinase activity involved in innate immune response
GO:0039503 suppression by virus of host innate immune response
GO:0052170 negative regulation by symbiont of host innate immune response
GO:0052166 positive regulation by symbiont of host innate immune response
GO:0052167 modulation by symbiont of host innate immune response
GO:0052390 induction by symbiont of host innate immune response
GO:0052305 positive regulation of innate immune response in other organism
GO:0052309 negative regulation of innate immune response in other organism
GO:0002220 innate immune response activating cell surface receptor signaling pathway
GO:0038078 MAP kinase phosphatase activity involved in regulation of innate immune response
GO:0052157 modulation by symbiont of microbe-associated molecular pattern-induced host innate immune response
GO:0052169 pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response
GO:0052033 pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response
GO:0052034 negative regulation by symbiont of microbe-associated molecular pattern-induced host innate immune response
GO:0052382 induction by organism of innate immune response in other organism involved in symbiotic interaction
GO:0052306 modulation by organism of innate immune response in other organism involved in symbiotic interaction
GO:0052308 pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction
GO:0052258 negative regulation by organism of pathogen-associated molecular pattern-induced innate immune response of other organism involved in symbiotic interaction
GO:0052257 pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction
GO:0052296 modulation by organism of microbe-associated molecular pattern-induced innate immune response in other organism involved in symbiotic interaction
GO:0035663 Toll-like receptor 2 binding
GO:0035662 Toll-like receptor 4 binding
GO:0090644 age-related resistance
GO:0035325 Toll-like receptor binding
GO:0039699 viral mRNA cap methylation
GO:0046696 lipopolysaccharide receptor complex
GO:0002224 toll-like receptor signaling pathway
GO:0039539 suppression by virus of host MDA-5 signaling pathway
GO:0039538 suppression by virus of host RIG-I signaling pathway
GO:0039537 suppression by virus of host viral-induced cytoplasmic pattern recognition receptor signaling pathway
GO:0039560 suppression by virus of host IRF9 activity
GO:1990231 STING complex
GO:0002755 MyD88-dependent toll-like receptor signaling pathway
GO:0002756 MyD88-independent toll-like receptor signaling pathway
GO:0035669 TRAM-dependent toll-like receptor 4 signaling pathway
GO:0035668 TRAM-dependent toll-like receptor signaling pathway
GO:0035667 TRIF-dependent toll-like receptor 4 signaling pathway
GO:0035666 TRIF-dependent toll-like receptor signaling pathway
GO:0035661 MyD88-dependent toll-like receptor 2 signaling pathway
GO:0035660 MyD88-dependent toll-like receptor 4 signaling pathway
GO:0035665 TIRAP-dependent toll-like receptor 4 signaling pathway
GO:0035664 TIRAP-dependent toll-like receptor signaling pathway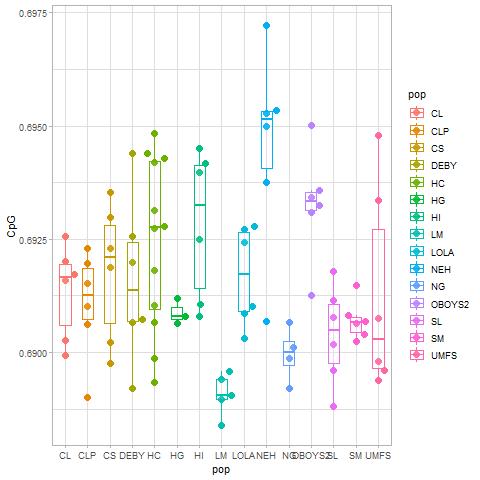


http://gannet.fish.washington.edu/Atumefaciens/20190225_cpg_oe/ID_CpG_labelled_all