We discussed this on the call, in addition to the stretching and scaling done above, we also decided on rotating the data after this step, using rgl::rotate3d. The angle needs to be a random sample from -pi to pi, and the unit vector needs to be random. Like generate a sequence c(random(-1,1), random(-1,1), random(-1,1)), where random(-1,1) is a random value from -1 to 1. Then normalize the vector to be of unit norm.
This came up as I was working on #37. It may be useful to include a function for generating "new" samples by adding noise to existing samples, then running the newly generated sample through the feature extraction pipeline. This step should happen just after a detrend (or perhaps a bandpass?) in the usual accel/gyro feature extraction pipeline.
Why generate new, fake samples? It's often the case that researchers are working with small amounts of data (from clinical studies) or there is a huge class imbalance between certain demographics (old/young, parkinsons/contol, ...) as we saw in mPower. To achieve better model accuracy, it's useful to augment the data by generating new samples. If we simply duplicate the data, it's oversampling. If we add noise, rotations, and/or modify the magnitude of the signal, it's data augmentation.
For example, here's the x axis of some accelerometer data: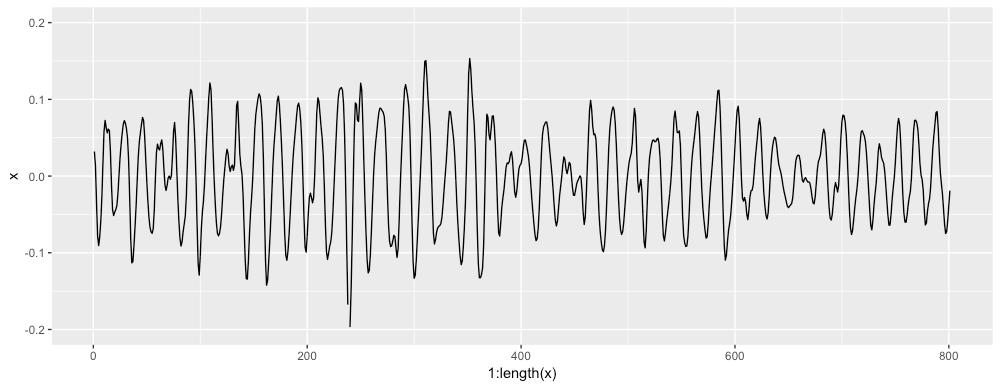
And here is the same signal with some noise added and magnitude modifications: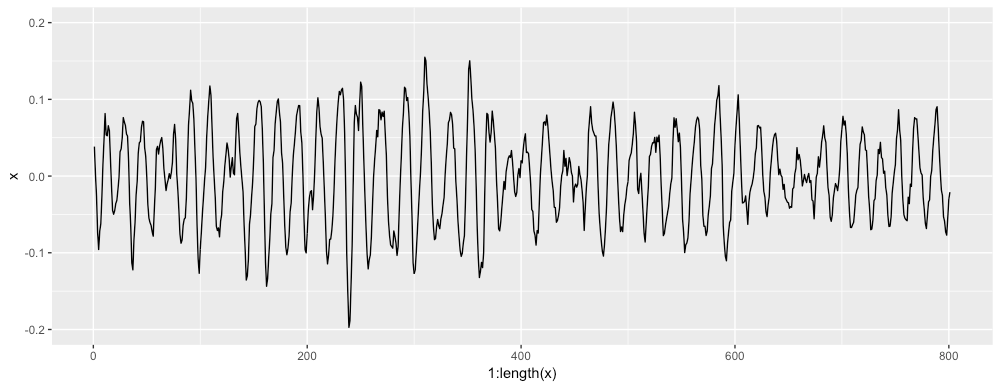
I'm using this code to transform the signal (sensor_data has columns x, y, z):