Thanks for sharing. Let us know if you need any help putting together the Julia version.
Closed Emmanuel-R8 closed 10 months ago
Thanks for sharing. Let us know if you need any help putting together the Julia version.
Calling on the offer.
The post includes an example which basically replicates a multimodal 2D distribution from a PyTorch library. See [https://torchdyn.readthedocs.io/en/latest/tutorials/07a_continuous_normalizing_flows.html]().
The following is an attempt at a Julia translation using FFJORD. Despite my best efforts, it bombs out at the end of the attached code before finishing a single iteration. All the same whether using 1 or 2 dimensions. . Julia = 1.5.2, DiffEqFlux = 1.24, Flux = 0.11.1
Any advice?
using Random, Distributions, Plots, LinearAlgebra
n_samples = 1 << 9
n_gaussians = 6
n_dims = 2
# Translated from Torchdyn ToyDataset()
# :param n_samples::int : number of data points in the generated dataset
# :param n_gaussians::int : number of gaussians distributions placed on the circle of radius `radius`
# :param radius:: int : radius of the circle on which the distributions lie
# :param std_gaussians::int : standard deviation of the gaussians.
# :param noise:: float32 : standard deviation of noise magnitude added to each data point
function generate_gaussians(; n_dims = n_dims, n_samples=100, n_gaussians=7,
radius=1.f0, std_gaussians=0.2f0, noise=0.001f0)
x = zeros(Float32, n_dims, n_samples * n_gaussians)
y = zeros(Float32, n_samples * n_gaussians)
incremental_angle = 2 * π / n_gaussians
dist_gaussian = MvNormal(n_dims, sqrt(std_gaussians))
if n_dims > 2
dist_noise = MvNormal(n_dims - 2, sqrt(noise))
end
current_angle = 0.f0
for i ∈ 1:n_gaussians
current_loc = zeros(Float32, n_dims, 1)
if n_dims >= 1
current_loc[1] = radius * cos(current_angle)
end
if n_dims >= 2
current_loc[2] = radius * sin(current_angle)
end
x[1:n_dims, (i-1)*n_samples+1:i*n_samples] = current_loc[1:n_dims] .+ rand(dist_gaussian, n_samples)
if n_dims > 2
x[1:n_dims-2, (i-1)*n_samples+1:i*n_samples] = rand(noise, n_samples)
end
y[ (i-1)*n_samples+1:i*n_samples] = Float32(i) .* ones(Float32, n_samples)
current_angle = current_angle + incremental_angle
end
return x, y
end
X, Y = generate_gaussians(; n_samples = n_samples ÷ n_gaussians,
n_gaussians = n_gaussians,
radius = 4.0f0,
std_gaussians = 0.5f0)
X = (X .- mean(X)) ./ std(X)
X_SIZE = size(X)[2]
# We will continue onward using the GR backend
gr()
if n_dims == 1
histogram(X[1, :], title = "Sample from the true density")
else
scatter(X[1, :], X[2, :], title = "Sample from the true density", markershape=:x, markersize=2)
end
using DiffEqFlux, Optim, OrdinaryDiffEq, Zygote, Flux
f = Chain(Dense(n_dims, 16 * n_dims, tanh),
Dense(16 * n_dims, 16 * n_dims, tanh),
Dense(16 * n_dims, 16 * n_dims, tanh),
Dense(16 * n_dims, n_dims, tanh))
t_span = (0., 1.)
cnf_ffjord = FFJORD(f, t_span, Tsit5(), basedist = MvNormal(n_dims, 1.), monte_carlo = true)
function loss_adjoint_orig(θ)
logpx = cnf_ffjord(X, θ)[1]
return -mean(logpx)[1]
end
# callback function to observe training
cb = function()
global iter += 1
println("Iteration $iter")
end
# Train using the ADAM optimizer
iter = 0
res1 = DiffEqFlux.sciml_train(
loss_adjoint_orig,
cnf_ffjord.p,
ADAM(0.1),
cb = cb,
maxiters = 10)
MethodError: no method matching (::var"#11#12")(::Array{Float32,1}, ::Float32)
Stacktrace:
[1] macro expansion at /home/emmanuel/.julia/packages/DiffEqFlux/8UHw5/src/train.jl:123 [inlined]
[2] macro expansion at /home/emmanuel/.julia/packages/ProgressLogging/BBN0b/src/ProgressLogging.jl:328 [inlined]
[3] (::DiffEqFlux.var"#73#78"{var"#11#12",Int64,Bool,Bool,typeof(loss_adjoint_orig),Array{Float32,1},Params})() at /home/emmanuel/.julia/packages/DiffEqFlux/8UHw5/src/train.jl:64
[4] with_logstate(::Function, ::Any) at ./logging.jl:408
[5] with_logger at ./logging.jl:514 [inlined]
[6] maybe_with_logger(::DiffEqFlux.var"#73#78"{var"#11#12",Int64,Bool,Bool,typeof(loss_adjoint_orig),Array{Float32,1},Params}, ::LoggingExtras.TeeLogger{Tuple{LoggingExtras.EarlyFilteredLogger{ConsoleProgressMonitor.ProgressLogger,DiffEqFlux.var"#68#70"},LoggingExtras.EarlyFilteredLogger{Base.CoreLogging.SimpleLogger,DiffEqFlux.var"#69#71"}}}) at /home/emmanuel/.julia/packages/DiffEqFlux/8UHw5/src/train.jl:39
[7] sciml_train(::Function, ::Array{Float32,1}, ::ADAM, ::Base.Iterators.Cycle{Tuple{DiffEqFlux.NullData}}; cb::Function, maxiters::Int64, progress::Bool, save_best::Bool) at /home/emmanuel/.julia/packages/DiffEqFlux/8UHw5/src/train.jl:63
[8] top-level scope at In[124]:3
[9] include_string(::Function, ::Module, ::String, ::String) at ./loading.jl:1091
[10] execute_code(::String, ::String) at /home/emmanuel/.julia/packages/IJulia/rWZ9e/src/execute_request.jl:27
[11] execute_request(::ZMQ.Socket, ::IJulia.Msg) at /home/emmanuel/.julia/packages/IJulia/rWZ9e/src/execute_request.jl:86
[12] #invokelatest#1 at ./essentials.jl:710 [inlined]
[13] invokelatest at ./essentials.jl:709 [inlined]
[14] eventloop(::ZMQ.Socket) at /home/emmanuel/.julia/packages/IJulia/rWZ9e/src/eventloop.jl:8
[15] (::IJulia.var"#15#18")() at ./task.jl:356
The callback needs to allow 2 args, just make it be
cb = function(x,l)
global iter += 1
println("Iteration $iter")
endand it should work
Did the trick. Thanks a lot.
I never thought about looking there to find the error since I used the Augmented NODE callback [https://diffeqflux.sciml.ai/dev/examples/augmented_neural_ode/#Callback-Function-1]() which has no parameters.
@Emmanuel-R8 the reason for that is sciml_train and Flux.train! have different requirements for the callback function. The augmented NODE example uses the latter.
Thanks Avik
I used this callback because the CNF example uses a callback ("cb=cb") which is not defined. So I went sniffing around.
When I have a bit of time, I'll work on a PR to improve the docs with the benefit of this discussion.
Despite playing with the code in ffjord.jl code for a few hours, I still find myself unable to fully use the model. I have a 2D model cnf_ffjord and a set of trained parameters res1. I want to be able to do several things:
cnf_ffjord([x, y], res1.minimizer; monte_carlo=false) to do that. Running that over a range of values gives a min of -9 and max of 29. Clearly I don't pass the right parameters or I don' t understand what cnf_ffjord does.@DhairyaLGandhi could we get some help here?
@DhairyaLGandhi
Anything I can provide to assist?
Oh actually it's probably @d-netto that would be able to comment on that function, sorry.
You'd also have to return a bool from the callback as sciml_train expects. I was able to train the model by eliminating the callback, since it isn't doing anything at the moment. One other quick change I made was
f = Chain(Dense(n_dims, 16 * n_dims, tanh),
Dense(16 * n_dims, 16 * n_dims, tanh),
Dense(16 * n_dims, 16 * n_dims, tanh),
Dense(16 * n_dims, n_dims, tanh)) |> Flux.f64 # <- note thisThe issue is convergence (I'll ignore speed for the moment). By plotting intermediate steps, I discovered it is an absolute dog's breakfast.
I am trying to learn 6 normal distributions located on an hexagon:

Here are 2 series of plots each showing about 20 learning steps using ADAM. The former is with ADAM(0.1). The latter is with ADAM(0.02)


I now understand why trying to understand the training results was a road to nowhere. In the former, the plot is lost within 2 iterations. In the latter, it is a bit better but still not able to learn 6 modes, unlike the Torchdyn library (plot from my post):
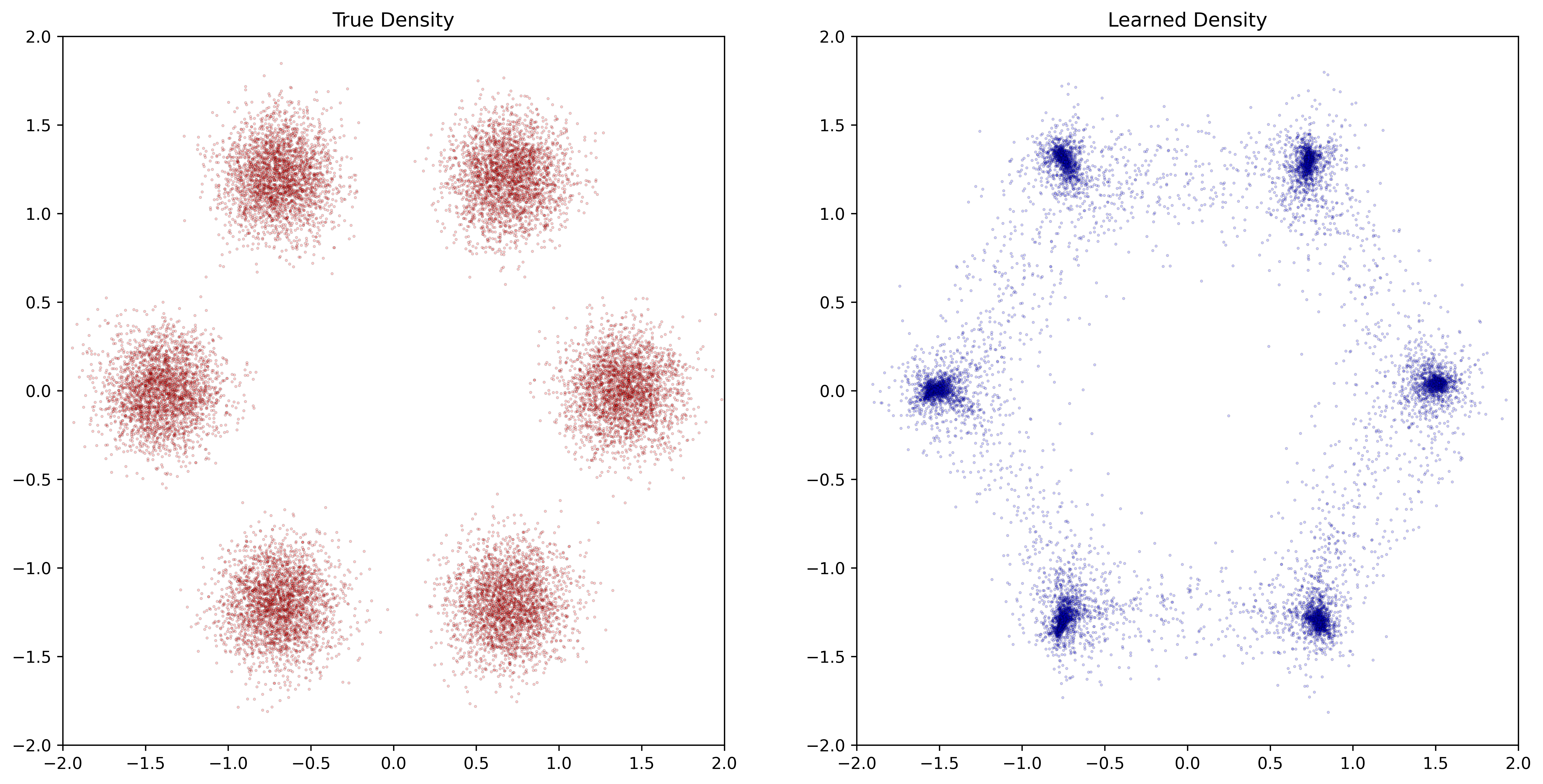 .
.
Any advice on how to improve convergence (get any at all)?
@d-netto let's talk about this.
# Code for Sampling
pz = cnf_ffjord.basedist
Z_samples = rand(pz, 512) |> gpu
sense = InterpolatingAdjoint()
ffjord_ = (u, p, t) -> DiffEqFlux.ffjord(u, p, t, cnf_ffjord.re, e, false, false)
e = cu(randn(eltype(X), size(Z_samples)))
_z = Zygote.@ignore similar(X, 1, size(Z_samples, 2))
Zygote.@ignore fill!(_z, 0.0f0)
prob = ODEProblem{false}(ffjord_, vcat(Z_samples, _z), (1.0, 0.0), res1.minimizer)
x_gen = solve(prob, cnf_ffjord.args...; sensealg = sense, cnf_ffjord.kwargs...)[1:end-1, :, end]
scatter(x_gen[1, :], x_gen[2, :], title = "Testing", markershape=:x, markersize=2)@Emmanuel-R8 There are a couple of suggestions I would make:
A good starting point is to always use the hyperparameters (+ model architecture) from a code that is known to work.
 The previous plot was of something else. This is the correct version.
The previous plot was of something else. This is the correct version.
Thanks.
I don' t have a GPU, and I haven' t yet succeeded to make Colab comply with my wishes. I'll need a few hours to check on my side.
My laptop is still churning along.
I am sharing where I am at on a Colab notebook. Not asking for review, but it took me a while to get that working, so I assume it will be useful to some.
https://colab.research.google.com/drive/1xbeWxgwrToZBYg8KxUOPjraoLrHrXbEJ?usp=sharing
Wrong link. This is the one:
https://colab.research.google.com/drive/15mBDXEfEv7Yg1t9Euw7CMFvKyQyNdh1f?usp=sharing
That's 100 iterations with 2e-3. A lot better. Thanks for the guidance.

This is a shameless plug for a post on Normalising flows and Neural ODEs. Short on Julia code for the moment but this is in the works.
I wanted to make any parts of the post available to be pilfered if anything can be used in the documentation. Otherwise comments welcome.
https://emmanuel-r8.github.io/post/2020/09/11/normalising-flows/