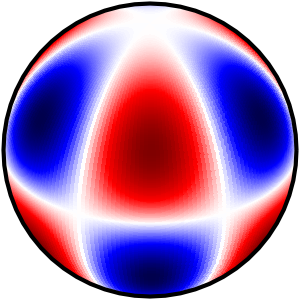
If you change your longitudes to be -pi -> pi rather than 0 -> 2*pi, contourf works as expected. Weird that it would work differently in the two different methods depending on the longitudes.
This is the change I made:
lon = np.linspace(0,2*np.pi,200) - np.pi








Description
I'm trying to draw spherical harmonic contours on an orthographic projection of a sphere. I see the correct contours when calling contour(), but many are not filled by contourf(). I think this is a bug, perhaps related to Issue 1024 (but I couldn't view their example figures).
Code to reproduce
Traceback
No error reported.
Full environment definition
### Operating system OS X 10.10.5 ### Cartopy version 0.16.0 ### conda list ``` # packages in environment at /Users/keatonb/anaconda: # # Name Version Build Channel _license 1.1 py27_0