/run standalone
Closed GNiendorf closed 8 months ago
/run standalone
The PR was built and ran successfully in standalone mode. Here are some of the comparison plots.
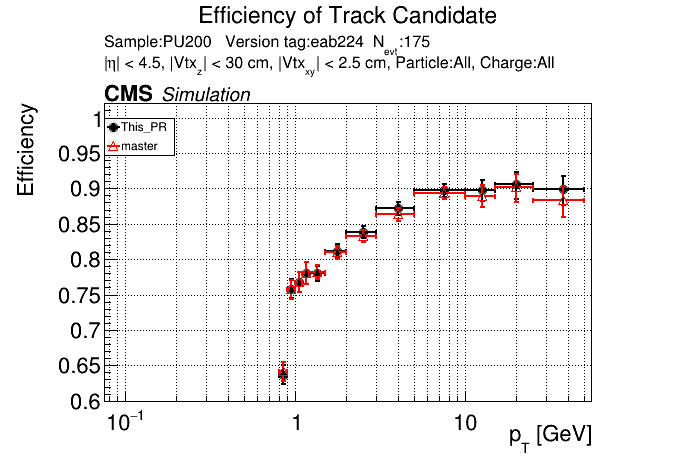 |
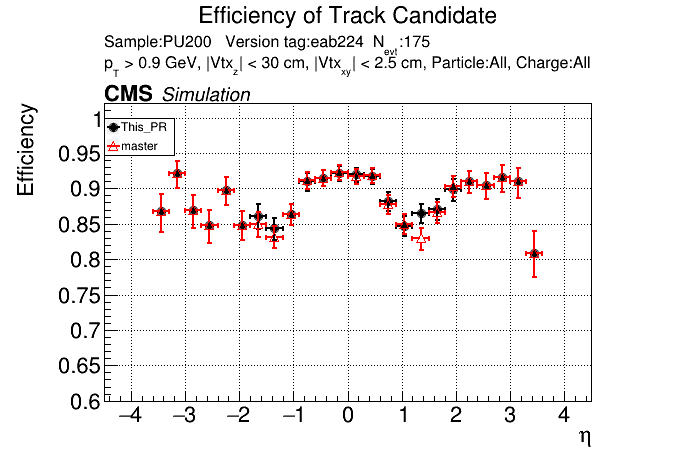 |
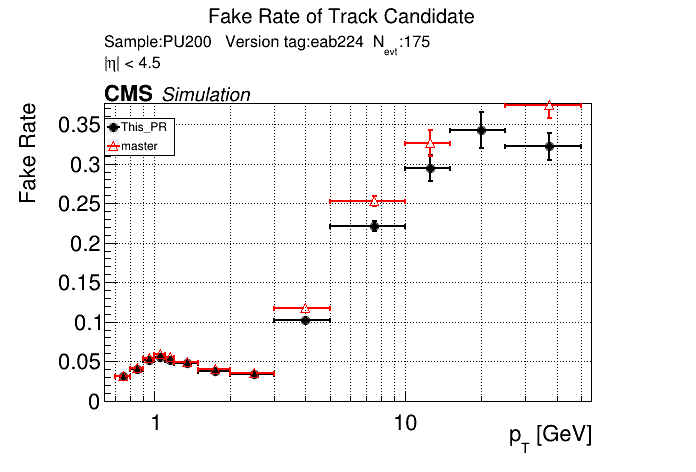 |
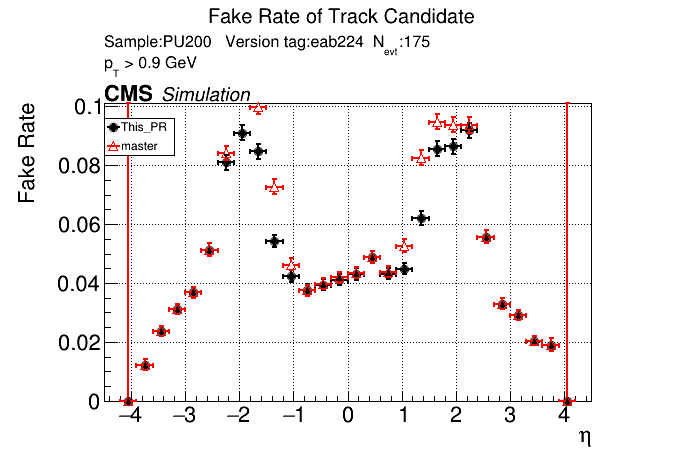 |
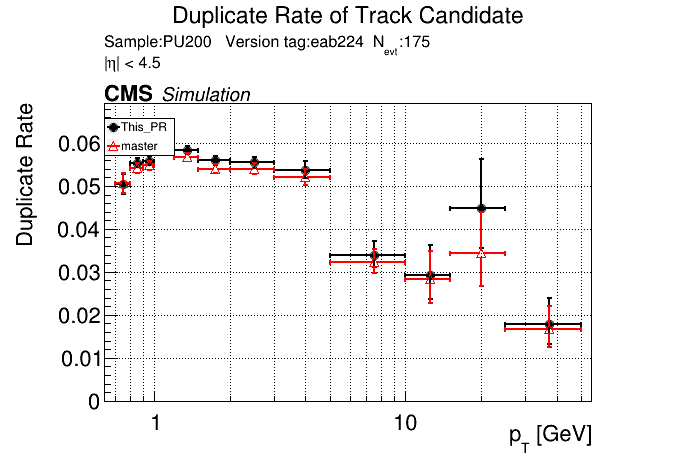 |
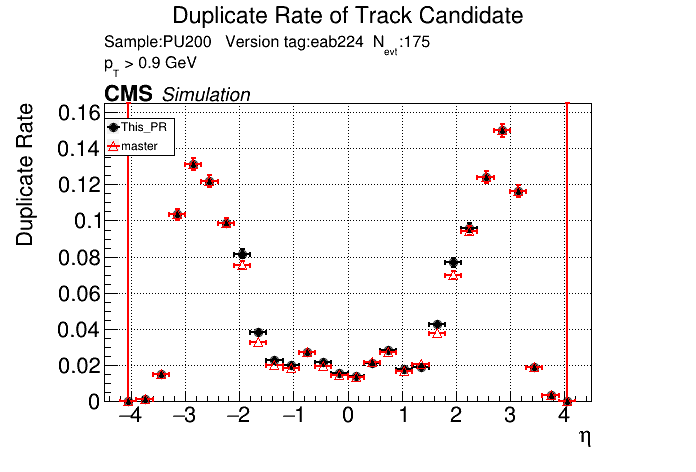 |
The full set of validation and comparison plots can be found here.
I think there is an error on the master branch. The code expects that the centroid values are attached to the endcap orientation file, see below.
But if you look at the file the master branch is using,
It doesn't have the centroid values appended and so it is filling those values with 0's. The centroid phi's are used in the endcap map, so those values are used by the algorithm. @VourMa I'll fix this in this PR.
/run cmssw
There was a problem while building and running with CMSSW. The logs can be found here.
There was a problem while building and running with CMSSW. The logs can be found here.
Looks like the master branch failed? @ariostas
Huh that's weird. I'm just gonna rerun the job and see if it works.
There was a problem while building and running with CMSSW. The logs can be found here.
I'm not sure what's happening. I'll look into it.
Oh it's because since https://github.com/SegmentLinking/cmssw/pull/16 CMSSW uses the LST headers, including Constants.h. So, since some numbers changed there, it would have to be recompiled. I'll implement that since it might be good to always recompile the LST part between PRs and master runs. I'll let you know when it's done.
was there a good reason to put new txt files in data/output ? why not directly in data/ ?
output is not a particularly meaningful additional layer.
was there a good reason to put new txt files in
data/output? why not directly indata/?outputis not a particularly meaningful additional layer.
also, with 2M lines proposed to be added, perhaps it's time to think of migrating to binary files from plain text for numbers.
was there a good reason to put new txt files in
data/output? why not directly indata/?outputis not a particularly meaningful additional layer.
This and other stuff will get cleaned up in the next commit. Still a draft PR for now.
All CSV, This PR: Here All CSV, but with old endcap file (i.e. with bug above): Here Comparison plots: Here
I'm just showing in the plots linked above that almost all of the performance differences we see are coming from fixing the bug in #375. The difference in switching from hit-based to CSV is small and only present in a couple of bins, as I've shown before.
I removed all of the old geometry files in the /data/ folder and moved the new CSV files from /output/ to /OT800_IT615_pt0.8/ with a new README file in /data/.
I'll look at @slava77's request now of moving from .txt files to binary files.
The PR was built and ran successfully with CMSSW. Here are some plots.
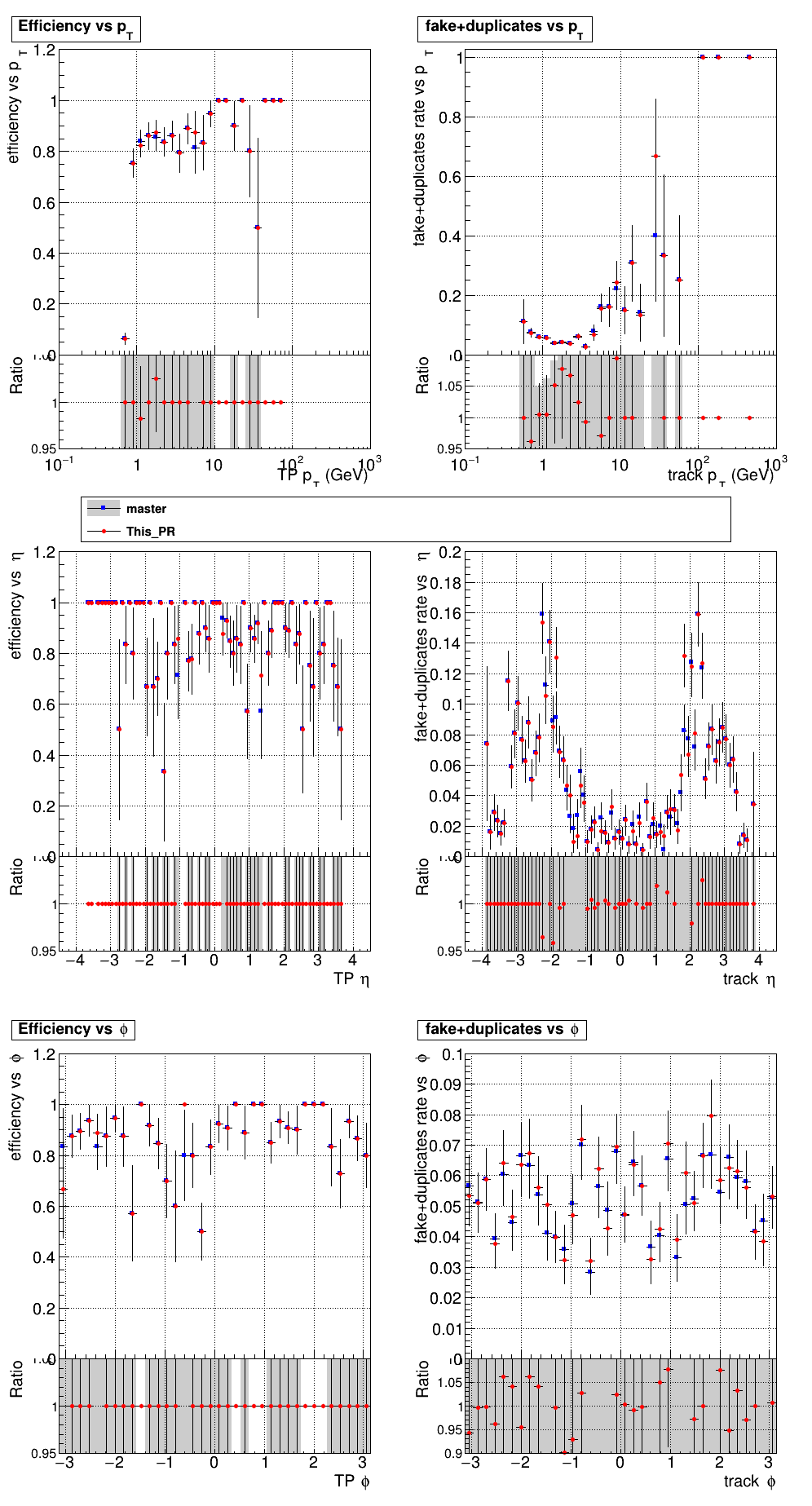 |
The full set of validation and comparison plots can be found here.
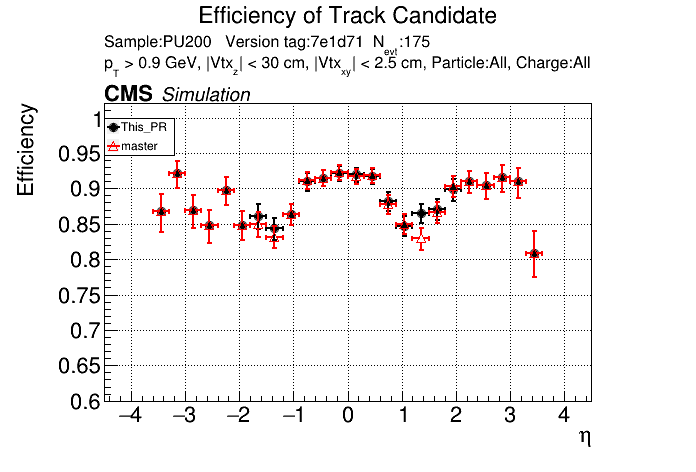
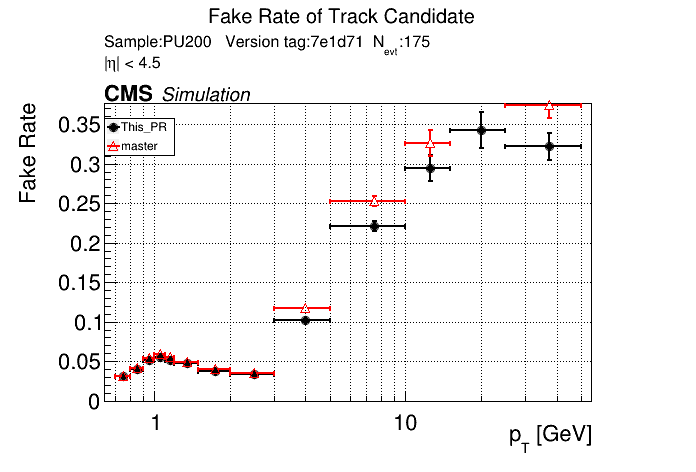
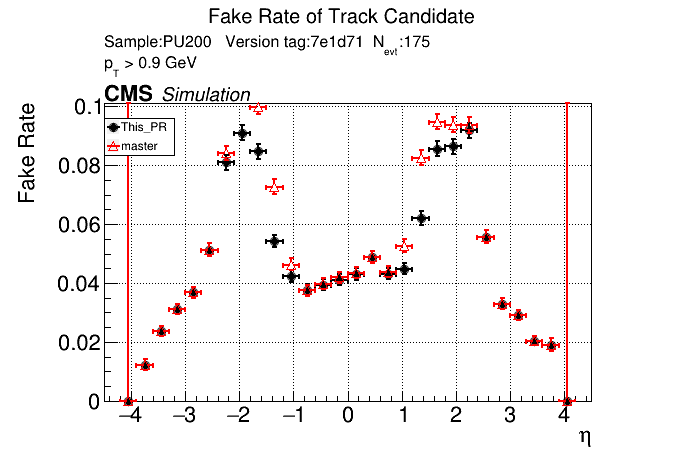
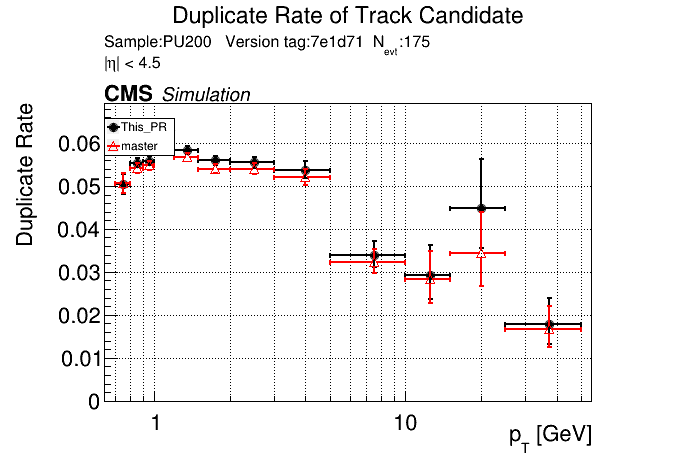
/run standalone
The PR was built and ran successfully in standalone mode. Here are some of the comparison plots.
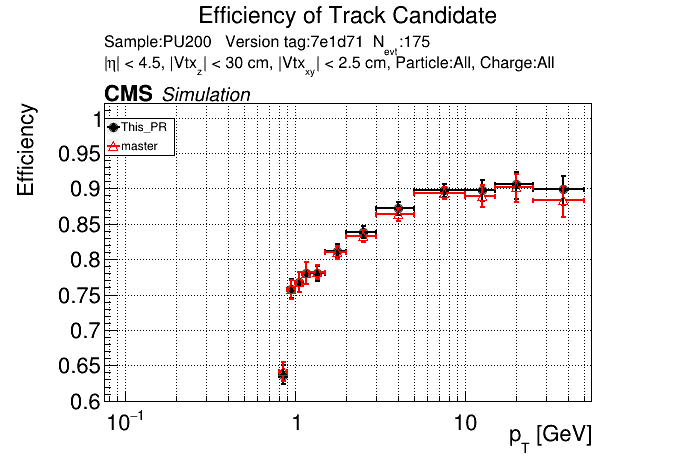 |
 |
 |
 |
 |
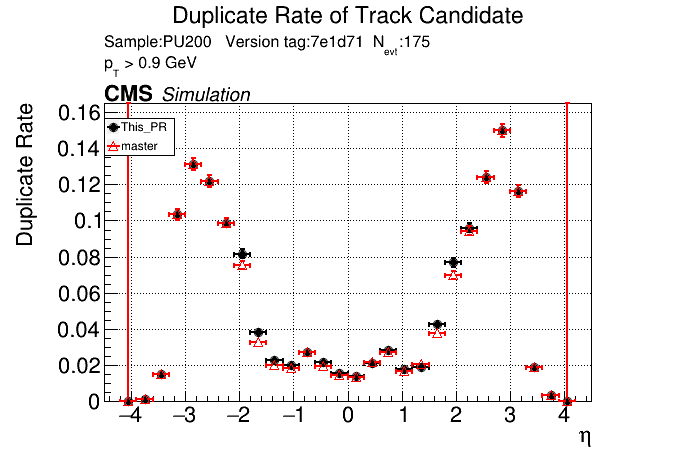 |
The full set of validation and comparison plots can be found here.
/run standalone
The PR was built and ran successfully in standalone mode. Here are some of the comparison plots.
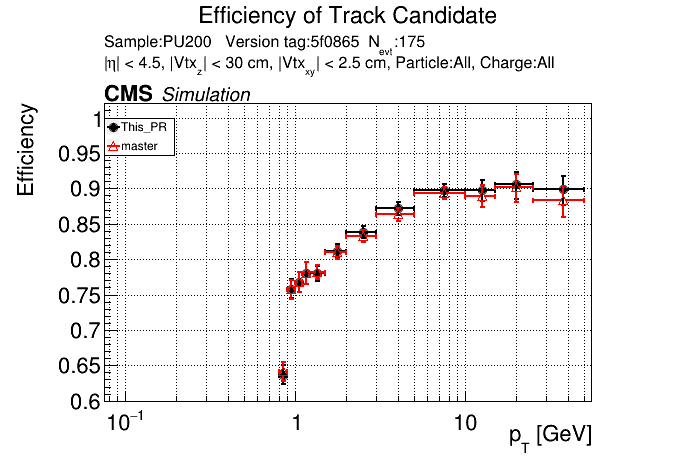 |
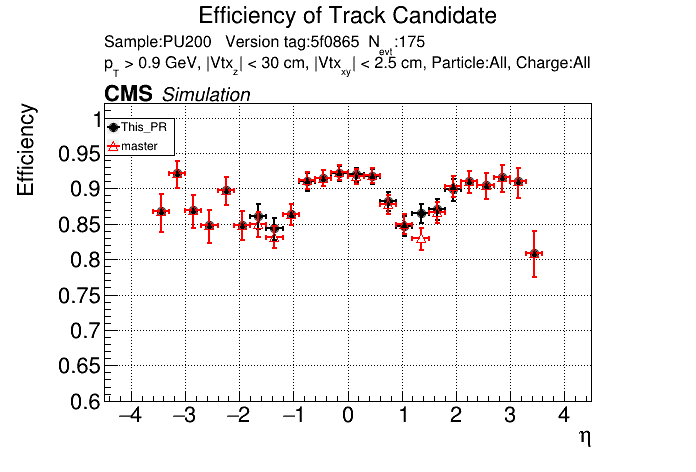 |
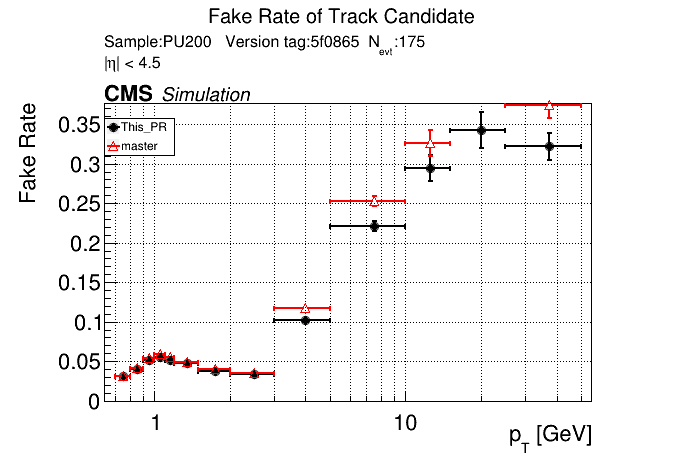 |
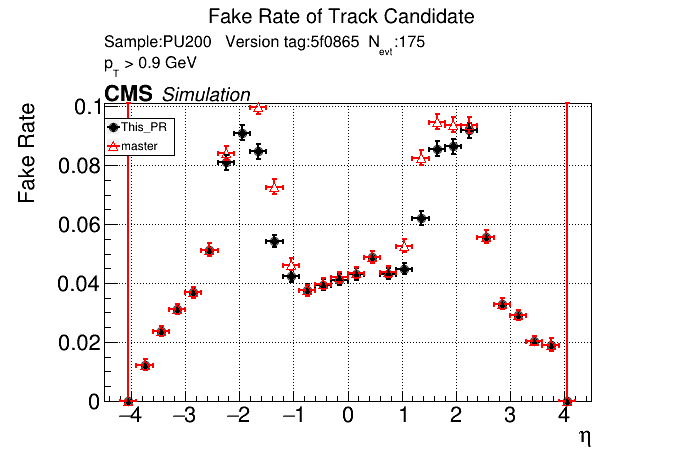 |
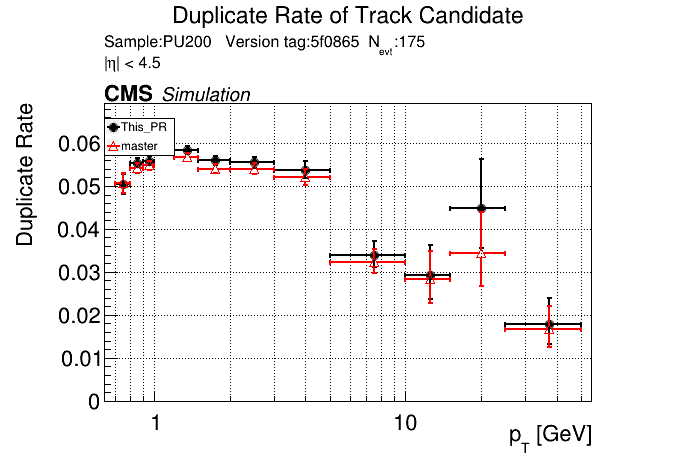 |
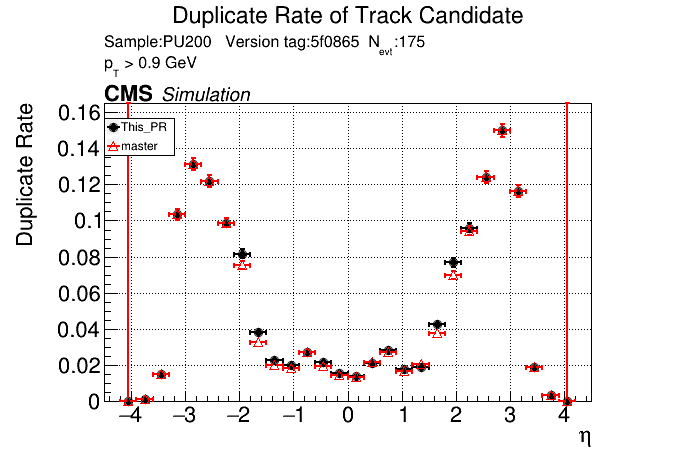 |
The full set of validation and comparison plots can be found here.
/run cmssw CMSSW_13_3_0_pre3_LST_X
Just making sure that the CMSSW workflow works well for 100 events.
Also, just leaving an extra reminder to squash and merge since there are some very large intermediate diffs.
The PR was built and ran successfully with CMSSW. Here are some plots.
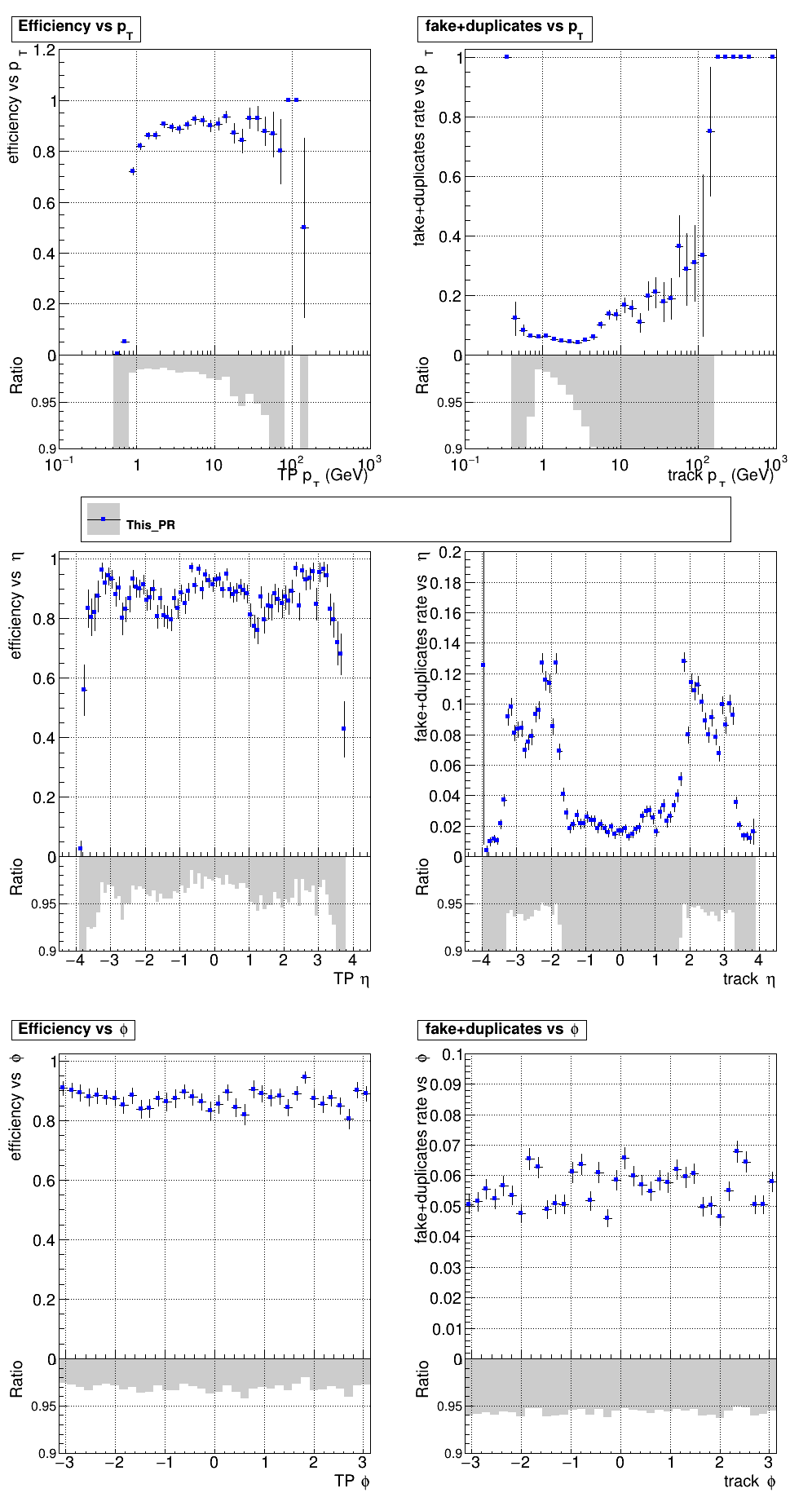 |
The full set of validation and comparison plots can be found here.
/run cmssw CMSSW_13_3_0_pre3_LST_X
Just making sure that the CMSSW workflow works well for 100 events.
Also, just leaving an extra reminder to squash and merge since there are some very large intermediate diffs.
Did you get rid of the comparison to master? Or is that in a different command.
There was a problem while building and running with CMSSW. The logs can be found here.
Did you get rid of the comparison to master? Or is that in a different command.
Now everything is switched to CMSSW_14_1_0_pre0, so I had to specify the older branch CMSSW_13_3_0_pre3_LST_X. When running with a non-default branch it doesn't run comparisons with master since there might be many things that changed so pretty much everything would have to be done again.
@GNiendorf While I am starting to look into the PR, could you please update the PR description with the actual changes that go into this PR? It is a bit hard to understand when there are 53 files changed and issues and fixes are mentioned in a incoherent way across O(30) comments. Thank you.
Summary of changes @VourMa:
Thanks, this looks good as well. Apart from the comments, I have a couple of general questions:
- What is the plan on replacing the
.txtwith binary files? Is it something for this PR?- My very naive expectation would be no changes in physics performance, so I can't say it is clear to me why validation plots show differences. Could you please explain?
We talked about binary files in one of the weekly meetings and came to the conclusion that it should be left for a different PR in the future. Also for number 2, the performance changes are coming from the bug I fixed in #375, where the endcap maps expected that the centroid phi position was being loaded but it wasn't, so those values were all being set to 0's.
/run standalone /run cmssw
The PR was built and ran successfully in standalone mode. Here are some of the comparison plots.
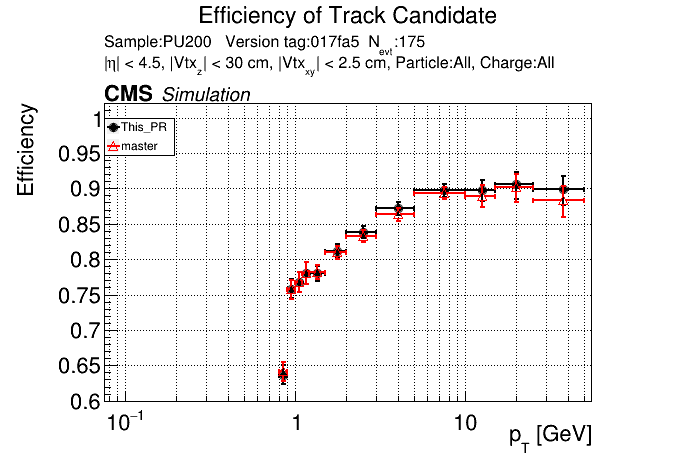 |
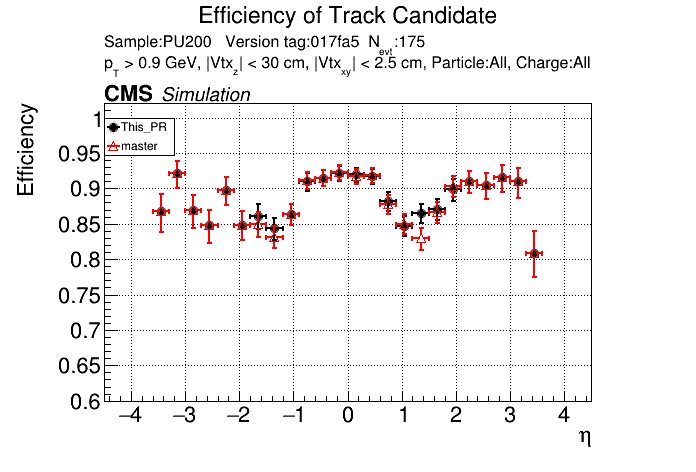 |
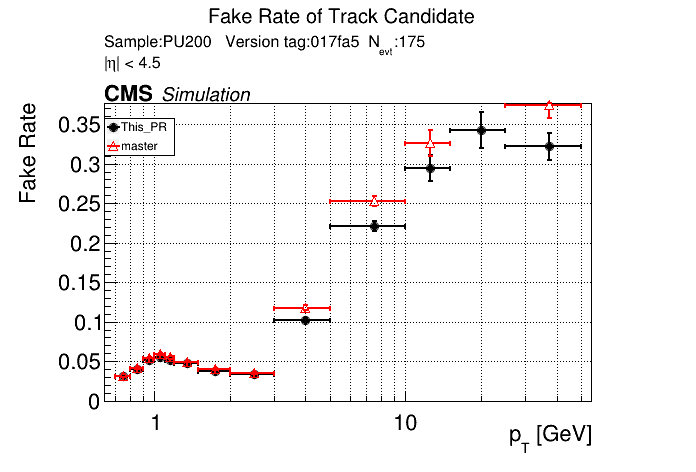 |
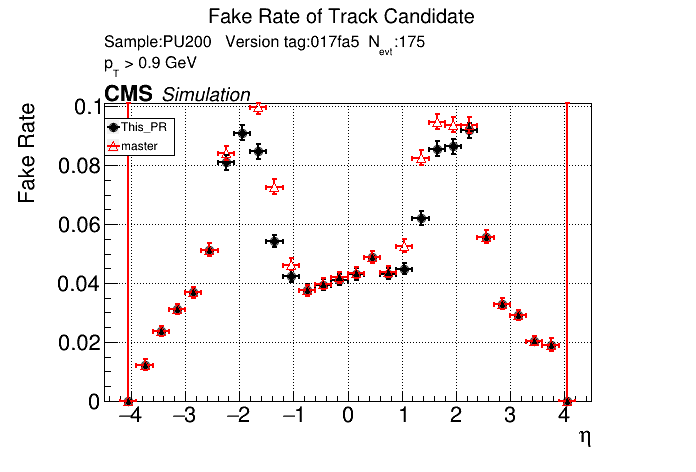 |
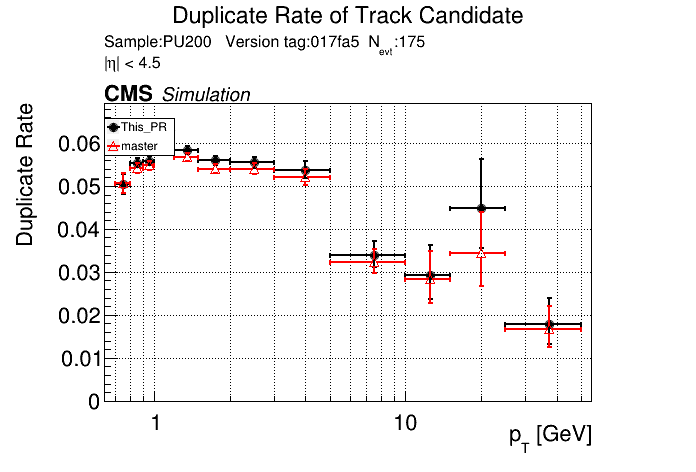 |
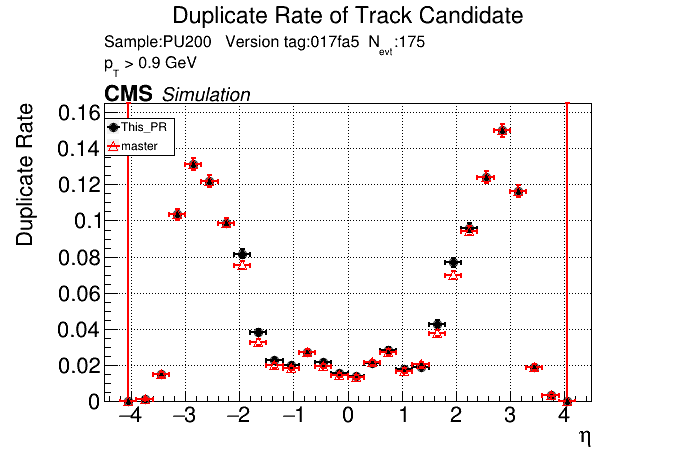 |
The full set of validation and comparison plots can be found here.
There was a problem while building and running with CMSSW. The logs can be found here.
There was a problem while building and running with CMSSW. The logs can be found here.
A fatal system signal has occurred: segmentation violation ./run.sh: line 76: 4133 Segmentation fault (core dumped) cmsRun step3_RAW2DIGI_RECO_VALIDATION_DQM.py
I'm not sure why there's a segfault. I'll take a look.
@VourMa I thought there was another commit I had to make, but it looks like this PR is ready to be merged in as well (unless I missed something).
This PR is associated with the corresponding PR on LSTGeometry: https://github.com/SegmentLinking/LSTGeometry/pull/10