The script for generating the above dataset is attached:
import MEArec as mr
import numpy as np
import matplotlib.pylab as plt
import os
import scipy.io
####################################parameters######################################
probe = 'Neuropixels2-128'
fs = 20000
n_exc = 1
n_inh = 0
duration = 1 #seconds
filtering = True #filtering True/false
noise_type = 'uncorrelated' #uncorrelated/distance-correlated/far-neurons
####################################################################################
dir_full_templates = '/mearec/temp_datasets/templates300_40kHz_minampnull_'+ probe +'.h5'
dir_full_recordings = '/mearec/temp_datasets/recording_'+str(n_exc+n_inh) +'cells_' + '_noise10' +str(filtering)+'filter_'+str(fs)+'fs_'+probe +'.h5'
tempgen = mr.load_templates(dir_full_templates)
recording_params = mr.get_default_recordings_params()
# Set parameters
recording_params['spiketrains']['n_exc'] = n_exc
recording_params['spiketrains']['n_inh'] = n_inh
recording_params['spiketrains']['duration'] = duration
recording_params['spiketrains']['seed'] = 0
#recording_params['spiketrains']['ref_per'] = 20
recording_params['templates']['min_amp'] = 40
recording_params['templates']['max_amp'] = 300
recording_params['templates']['seed'] = 0
recording_params['recordings']['modulation'] = 'electrode'
recording_params['recordings']['noise_mode'] = noise_type
recording_params['recordings']['noise_level'] = 10
# use chunk options
recording_params['recordings']['chunk_conv_duration'] = 20
recording_params['recordings']['chunk_noise_duration'] = 20
recording_params['recordings']['chunk_filter_duration'] = 20
recording_params['recordings']['seed'] = 0
#filter settings
recording_params['recordings']['fs'] = fs
recording_params['recordings']['filter'] = filtering
recording_params['recordings']['filter_cutoff'] = [300, 6000] # highpass from 0
#seeds settings
recording_params['seeds']['spiketrains'] = 0
recording_params['seeds']['templates'] = 0
recording_params['seeds']['convolution'] = 0
recording_params['seeds']['noise'] = 0
recgen = mr.gen_recordings(templates=dir_full_templates, params=recording_params, verbose=True)
# save recording
mr.save_recording_generator(recgen, dir_full_recordings)

Hi there, I am using the MEArec and I found a problem: the filtering of MEArec seems not functioning well. It seems to me that the filtered signals are added on the non-filtered signal, while I expected the filtered signal (only) to be the output. In the source code here: https://github.com/alejoe91/MEArec/blob/2e83a35c386616830e08f799577b419a61a9f94b/MEArec/generators/recgensteps.py#L47
I see the
full_arr/args/kargsare updated tofull_arr + out_chunk.Because I check a case (in which I can put the settings below for you to reproduce this), and I found the variable
signals_filtgenerated by https://github.com/alejoe91/MEArec/blob/2e83a35c386616830e08f799577b419a61a9f94b/MEArec/tools.py#L2592 is not the same as the generated dataset's output. I see they are added with the raw recordings from the previous step. For example, in the above case, theful_arrbefore L47 is the raw recording ( the same assignalsin functionfilter_analog_signals) . The first 5 samples of ch0 are:and the
out_chunkis the filtered signal the same assignals_filtin functionfilter_analog_signals. The first 5 samples of ch0 are:And after updating, the
full_arris updated to:And the updated 'full_arr' is the output of mearec datasets. (i.e.
.recordings)And I plot the spectrum of the output recording using pwelch in matlab, the spectrum of the filtered data is: The roll-off is weird. BTW, I also plot the spectrum of
The roll-off is weird. BTW, I also plot the spectrum of  I also checked the spectrum with using unfiltered MEArec dataset + matlab filtfilt function, the spectrum is very similar to the spectrum of
I also checked the spectrum with using unfiltered MEArec dataset + matlab filtfilt function, the spectrum is very similar to the spectrum of 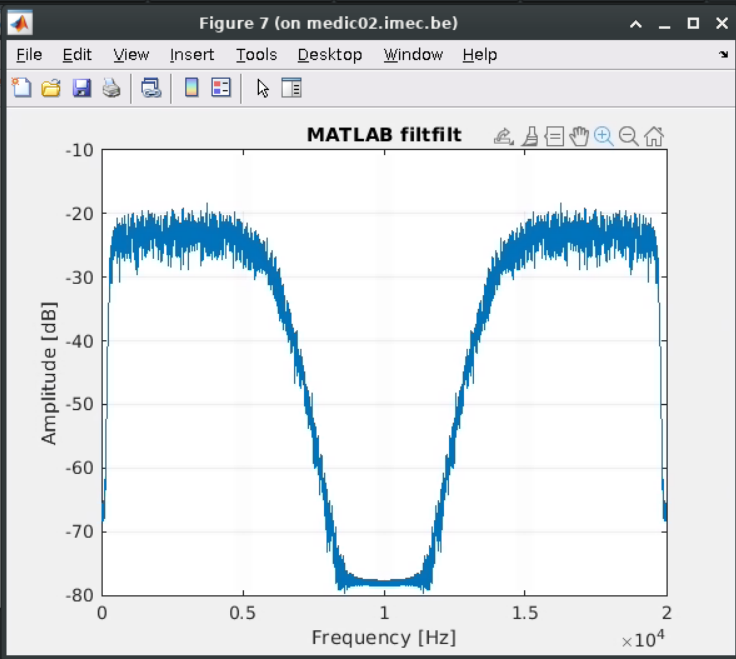
signals_filt, which seems correct:signals_filt.