Which terminal are you using? I'm guessing you are on Windows? Can you use the Powershell?
Closed justinjjlee closed 3 years ago
Which terminal are you using? I'm guessing you are on Windows? Can you use the Powershell?
I am using VScode and julia addon/interface.
@justinjoliver Are you asking for the stansummary output? That output also seems to contain these page make up characters. Is this VSCode running on Windows?
That is what I am trying to see. but the bits of string is just covering the entire command prompt for julia. Regardless of what I am querying, I can't see anything (not even what I am typing).

Yes - I am running VSCode running on Windows.
Can you send me the script you are trying to run?
Just ran a simple script on VSCode, including a call to read_summary(), but no problems on my system (macOS). Recently I helped someone else who was able to switch to Powershell (from the github bash shell I believe), which resolved thsis issue.
I'll check a bit if that would be feasible in VSCode.
Ok, just installed PowerShell on my system and then installed it in VSCode. If I then run a Julia script, pwsh seems to turn into the Julia REPL. Guess that's the closest I can come to emulate Windows on my system.
I'm assuming you can start a Julia REPL and execute non-Cmdstan scripts in that REPL?
Above you write "That is what I am trying to see". You mean the output of stansummary by calling read_summary(model)?
So, I can execute any operations. The issue is not related to the operation of Stan, but relates to what happens afterwards.
After I run the Stan model, my REPL is covered by the the text shown in the image - instead of command line starting with 'julia>'
FYI - I was running example bernoulli example.
Hi Justin,
Great, what I'm trying to find out is what step in executing a script causes the shell to go nuts. E.g. can you execute line by line below program, e.g. by copy-pasting into the REPL?
using CmdStan
bm = "
data {
int<lower=1> N;
int<lower=0,upper=1> y[N];
}
parameters {
real<lower=0,upper=1> theta;
}
model {
theta ~ beta(1,1);
y ~ bernoulli(theta);
}
";
data = Dict("N" => 10, "y" => [0, 1, 0, 1, 0, 0, 0, 0, 0, 1])
sm = Stanmodel(name="bernoulli", model=bm, printsummary=false);
data # Just to check if terminal is still ok
rc, samples, cnames = stan(sm, data, pwd(), CmdStanDir=CMDSTAN_HOME);
samples # Just checking again
if rc == 0
sum_df = read_summary(stanmodel)
draw_df = read_samples(
end
println()
sum_dfAh, got you. Hope I can be more helpful.
I have executed the program above. The phenomenon starts with the line,
rc, samples, cnames = stan(sm, data, pwd(), CmdStanDir=CMDSTAN_HOME);
As you can see from the image below.

Because the sample prints the long dataframe, I have cut the image and am showing the rest below,

Hope this helps, and thank you so much for the (very very timely) troubleshoot. I really appreciate it!
One more, hopefully quick test. Can you replace the lines after the data definition by:
...
data = Dict("N" => 10, "y" => [0, 1, 0, 1, 0, 0, 0, 0, 0, 1])
sm = Stanmodel(name="bernoulli", model=bm, printsummary=false)
sm |> display
rc, samples, cnames = stan(sm, data; summary=false, CmdStanDir=CMDSTAN_HOME);
nt = CmdStan.convert_a3d(samples, cnames, Val(:namedtuple))
mean(nt.theta, dims=1) |> displayIn above test I have explicitly disabled running stansummary (under the covers, part of stan()).
Have you ever tried something like:
m = rand(3,4,5)
m |> displayIf that works fine, we know for sure it is caused by something in CmdStan.jl because in the first run samples is a simple 3 dimensional array.
So, your first test, the phenomenon persists,
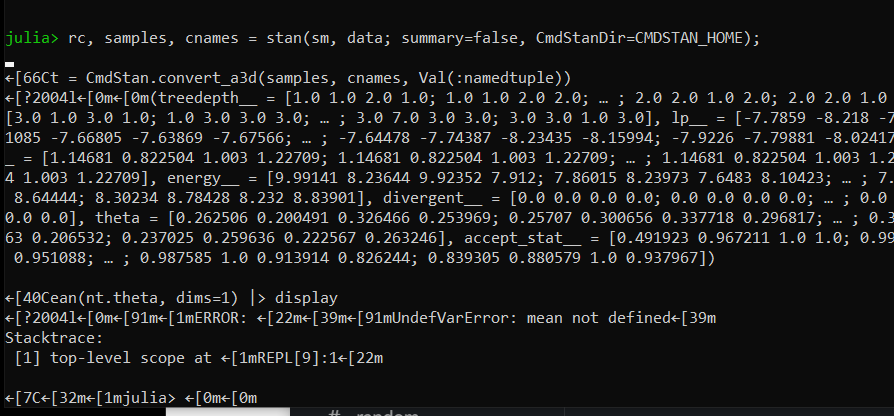
For your second test - it seems like it is causing any issue.

In your terminal (where you see the above output) I have a control. which shows the shells being used, in my case

If you click on what is showing Julia REPL you should see at least 2 shells. Which are those? If you click on the down arrow and select Select Default Shell, what are your options?
For the first: I only see 'Julia REPL' For the second: I see two: 'Command Prompt' and 'Windows PowerShell'
Can you select Windows PowerShell?
Not sure if you then have to restart VSCode. I noticed in the latest test script I forgot to include Statistics on the first line:
using CmdStan, Statistics
bm = "
data {
int<lower=1> N;
int<lower=0,upper=1> y[N];
}
parameters {
real<lower=0,upper=1> theta;
}
model {
theta ~ beta(1,1);
y ~ bernoulli(theta);
}
";
data = Dict("N" => 10, "y" => [0, 1, 0, 1, 0, 0, 0, 0, 0, 1])
sm = Stanmodel(name="bernoulli", model=bm, printsummary=false)
sm |> display
rc, samples, cnames = stan(sm, data; summary=false, CmdStanDir=CMDSTAN_HOME);
nt = CmdStan.convert_a3d(samples, cnames, Val(:namedtuple))
mean(nt.theta, dims=1) |> display
sm.command |> displayUpdate: But with using Statistics in there the output in your screenshot would be correct (except for the make-up characters). The NamedTuple starting with tree depth__ is correct.
What would also help me if you could send the output of both versioninfo() and ENV.
Did with windows powershell - same deal. Did included Statistics when I saw the functions.
My version info:
Julia Version 1.5.4
Commit 69fcb5745b (2021-03-11 19:13 UTC)
Platform Info:
OS: Windows (x86_64-w64-mingw32)
CPU: Intel(R) Xeon(R) CPU E3-1270 v5 @ 3.60GHz
WORD_SIZE: 64
LIBM: libopenlibm
LLVM: libLLVM-9.0.1 (ORCJIT, skylake)Environment:
JULIA_CMDSTAN_HOME = C:\cmdstan
JULIA_EDITOR = "C:\Users\Justin\AppData\Local\Programs\Microsoft VS Code\Code.exe"
JULIA_NUM_THREADS =ENV
Base.EnvDict with 51 entries:
"ALLUSERSPROFILE" => "C:\\ProgramData"
"APPDATA" => "C:\\Users\\Justin\\AppData\\Roa… "CHROME_CRASHPAD_PIPE_NAME" => "\\\\.\\pipe\\crashpad_14304_CZVAXLWQFBSVWBCI"
"CMDSTAN_HOME" => "C:\\cmdstan"
"COMMONPROGRAMFILES" => "C:\\Program Files\\Common Files"
"COMMONPROGRAMFILES(X86)" => "C:\\Program Files (x86)\\Common Files"
"COMMONPROGRAMW6432" => "C:\\Program Files\\Common Files"
"COMPUTERNAME" => "Justin"
"COMSPEC" => "C:\\windows\\system32\\cmd.exe"
"DRIVERDATA" => "C:\\Windows\\System32\\Drivers\\DriverData"
"FPS_BROWSER_APP_PROFILE_STRING" => "Internet Explorer"
"FPS_BROWSER_USER_PROFILE_STRING" => "Default"
"HOMEDRIVE" => "C:"
"HOMEPATH" => "\\Users\\Justin"
"JULIA_CMDSTAN_HOME" => "C:\\cmdstan"
"LOCALAPPDATA" => "C:\\Users\\Justin\\AppData\\Loc… "LOGONSERVER" => "\\\\Justin"
"NUMBER_OF_PROCESSORS" => "8"
"OPENBLAS_MAIN_FREE" => "1"
"ORIGINAL_XDG_CURRENT_DESKTOP" => "undefined"
"OS" => "Windows_NT"
"PATH" => "C:\\windows\\system32;C:\\windows;C:\\windows\\Syst… "PATHEXT" => ".COM;.EXE;.BAT;.CMD;.VBS;.VBE;.JS;.JSE;.WSF;.WSH;.M… "PROCESSOR_ARCHITECTURE" => "AMD64"
"PROCESSOR_IDENTIFIER" => "Intel64 Family 6 Model 94 Stepping 3, GenuineIntel"
"PROCESSOR_LEVEL" => "6"
"PROCESSOR_REVISION" => "5e03"
"PROGRAMDATA" => "C:\\ProgramData"
"PROGRAMFILES" => "C:\\Program Files"
"PROGRAMFILES(X86)" => "C:\\Program Files (x86)"
"PROGRAMW6432" => "C:\\Program Files"
"PSMODULEPATH" => "C:\\Program Files\\WindowsPowerShell\\Modules;C:\\w… "PUBLIC" => "C:\\Users\\Public"
"RTOOLS40_HOME" => "C:\\rtools40"
"SESSIONNAME" => "Console"
"SYSTEMDRIVE" => "C:"
"SYSTEMROOT" => "C:\\windows"
"TEMP" => "C:\\Users\\Justin\\AppData\\Local\\Temp"
"TMP" => "C:\\Users\\Justin\\AppData\\Local\\Temp"
"USERDOMAIN" => "Justin"
"USERDOMAIN_ROAMINGPROFILE" => "Justin"
"USERNAME" => "Justin"
"USERPROFILE" => "C:\\Users\\Justin"
"WINDIR" => "C:\\windows"
"JULIA_EDITOR" => "\"C:\\Users\\Justin\\AppData\\L… "JULIA_NUM_THREADS" => ""
"TERM_PROGRAM" => "vscode"
"TERM_PROGRAM_VERSION" => "1.54.3"
"LANG" => "en_US.UTF-8"
"COLORTERM" => "truecolor"Justin, when you restart VSCode and start a Terminal (not the Julia REPL, but e.g. using the New Terminal in the Terminal menu item, which terminal is shown?
Powershell
Hmmm, I'm running out of ammunition. As a last attempt, can you run a pwsh Terminal outside VSCode, start Julia there and the copy-and-paste the test script in there?
In either case, my next step will be to ask on Julia Discourse if someone else has ran into this issue.
Ok, some interesting responses on my question on Discourse.
It looks like externally spawned commands occasionally disable the handling of escape characters. And VSCode is directly spawning the Julia REPL, so my suggestion to switch to Powershell couldn't work I guess.
A very simple test from above request for help:
"I wanted to use the REPL shell mode (type ";" in the REPL,
shell> ls
20201105_12 20201106_11 Manifest.toml Project.toml io_caen movers test_proj tutorial
←[7C←[32m←[1mjulia> ←[0m←[0m"but the teminal mode in the julia repl gets screwed up. A sort of work around is to use the windows cmd.exe to indirectly call the desired program like this":
shell> cmd /c ls
20201105_12 20201106_11 Manifest.toml Project.toml io_caen movers test_proj tutorial
julia>Can you try above quick test?
If so, this is a bigger problem than calling stan(). I'm still confused why it doesn't happen earlier when I'm compiling by spawning make. Maybe because make doesn't produce output?
On going through the code in CmdStan.jl, I noticed this should have been fixed some time ago. The command line commands I'm using to run cmdstan is stored in sm.command created in above script.
On unix-like systems the commands should look like:
julia> sm.command
4-element Vector{Base.AbstractCmd}:
`./bernoulli sample num_samples=1000 num_warmup=1000 save_warmup=0 thin=1 adapt engaged=1 gamma=0.05 delta=0.8 kappa=0.75 t0=10.0 init_buffer=75 term_buffer=50 window=25 algorithm=hmc engine=nuts max_depth=10 metric=diag_e stepsize=1.0 stepsize_jitter=1.0 random seed=-1 id=1 data file= bernoulli_1.data.R output file= bernoulli_samples_1.csv refresh=100`
`./bernoulli sample num_samples=1000 num_warmup=1000 save_warmup=0 thin=1 adapt engaged=1 gamma=0.05 delta=0.8 kappa=0.75 t0=10.0 init_buffer=75 term_buffer=50 window=25 algorithm=hmc engine=nuts max_depth=10 metric=diag_e stepsize=1.0 stepsize_jitter=1.0 random seed=-1 id=2 data file= bernoulli_2.data.R output file= bernoulli_samples_2.csv refresh=100`
`./bernoulli sample num_samples=1000 num_warmup=1000 save_warmup=0 thin=1 adapt engaged=1 gamma=0.05 delta=0.8 kappa=0.75 t0=10.0 init_buffer=75 term_buffer=50 window=25 algorithm=hmc engine=nuts max_depth=10 metric=diag_e stepsize=1.0 stepsize_jitter=1.0 random seed=-1 id=3 data file= bernoulli_3.data.R output file= bernoulli_samples_3.csv refresh=100`
`./bernoulli sample num_samples=1000 num_warmup=1000 save_warmup=0 thin=1 adapt engaged=1 gamma=0.05 delta=0.8 kappa=0.75 t0=10.0 init_buffer=75 term_buffer=50 window=25 algorithm=hmc engine=nuts max_depth=10 metric=diag_e stepsize=1.0 stepsize_jitter=1.0 random seed=-1 id=4 data file= bernoulli_4.data.R output file= bernoulli_samples_4.csv refresh=100`On Windows these should look like:
julia> sm.command
4-element Vector{Base.AbstractCmd}:
`cmd /c ./bernoulli.exe sample num_samples=1000 num_warmup=1000 save_warmup=0 thin=1 adapt engaged=1 gamma=0.05 delta=0.8 kappa=0.75 t0=10.0 init_buffer=75 term_buffer=50 window=25 algorithm=hmc engine=nuts max_depth=10 metric=diag_e stepsize=1.0 stepsize_jitter=1.0 random seed=-1 id=1 data file= bernoulli_1.data.R output file= bernoulli_samples_1.csv refresh=100`
`cmd /c ./bernoulli.exe sample num_samples=1000 num_warmup=1000 save_warmup=0 thin=1 adapt engaged=1 gamma=0.05 delta=0.8 kappa=0.75 t0=10.0 init_buffer=75 term_buffer=50 window=25 algorithm=hmc engine=nuts max_depth=10 metric=diag_e stepsize=1.0 stepsize_jitter=1.0 random seed=-1 id=2 data file= bernoulli_2.data.R output file= bernoulli_samples_2.csv refresh=100`
`cmd /c ./bernoulli.exe sample num_samples=1000 num_warmup=1000 save_warmup=0 thin=1 adapt engaged=1 gamma=0.05 delta=0.8 kappa=0.75 t0=10.0 init_buffer=75 term_buffer=50 window=25 algorithm=hmc engine=nuts max_depth=10 metric=diag_e stepsize=1.0 stepsize_jitter=1.0 random seed=-1 id=3 data file= bernoulli_3.data.R output file= bernoulli_samples_3.csv refresh=100`
`cmd /c ./bernoulli.exe sample num_samples=1000 num_warmup=1000 save_warmup=0 thin=1 adapt engaged=1 gamma=0.05 delta=0.8 kappa=0.75 t0=10.0 init_buffer=75 term_buffer=50 window=25 algorithm=hmc engine=nuts max_depth=10 metric=diag_e stepsize=1.0 stepsize_jitter=1.0 random seed=-1 id=4 data file= bernoulli_4.data.R output file= bernoulli_samples_4.csv refresh=100`Would you mind also checking that on your system? Just type sm.command in the terminal. It might be garbled. You could also append sm.command |> display to the test script (I added that in the post from 3 days ago).
It's very unfortunate I can't test any of this myself. But it would be really nice if VSCode + StanJulia would work on Windows!
From Tim I understand the combination of Stan.jl (.e.g. StanSample.jl etc.) + VSCode + Windows work ok. I wonder if that is another solution for you:
######### StanSample Bernoulli example ###########
using StanSample
ProjDir = @__DIR__
bernoulli_model = "
data {
int<lower=1> N;
int<lower=0,upper=1> y[N];
}
parameters {
real<lower=0,upper=1> theta;
}
model {
theta ~ beta(1,1);
y ~ bernoulli(theta);
}
";
bernoulli_data = Dict("N" => 10, "y" => [0, 1, 0, 1, 0, 0, 0, 0, 0, 1])
# Keep tmpdir across multiple runs to prevent re-compilation
tmpdir = joinpath(@__DIR__, "tmp")
sm = SampleModel("bernoulli", bernoulli_model;
method = StanSample.Sample(
save_warmup=false, # Default
thin=1,
adapt = StanSample.Adapt(delta = 0.85)),
tmpdir = tmpdir,
);
rc = stan_sample(sm; data=bernoulli_data);
if success(rc)
nt = read_samples(sm)
display(nt)
endDefinitely still interested in above output, but I'm afraid fixing CmdStan + VSCode + Windows might take a bit longer. It really should be fixed in VSCode or the Julia extension for VSCode.
Hi,
I am using the latest version of CmdStan.jl an Stan - 2.26.
Seems like wrapper is at work, but I am having some interface issue. After the first run, my julia REPL gets 'shadowed' by what it seems like an output. I can't really see anything after the run and I can't get it to disappear (tried print, read lines, escape, etc...). See the image below.
Any idea how I can troubleshoot this?
Thanks in advance.