My personal preferences are:
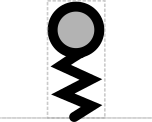
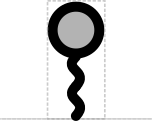
- Stem system: straight, wavy, sawtooth Nicely consistent with the typical usage of straight DNA backbone, wavy RNA, and sawtooth is a natural progression from wave.
- Tops:
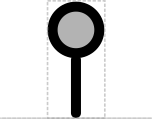
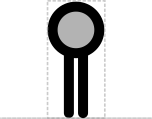
- Cut: circle Consistent with "map pins" and DeskGen usage
- X-ase: Binding cup Consistent with proposed Operator glyph and with protein language
- Stability: half-circle Sort of like a shield and frees up circle for Cut



 (from the Supplementary Materials of [Nielsen et al, *Science*, 2016](https://www.ncbi.nlm.nih.gov/pubmed/27034378))
(from the Supplementary Materials of [Nielsen et al, *Science*, 2016](https://www.ncbi.nlm.nih.gov/pubmed/27034378))
 (from the Supplementary Materials of [Nielsen et al, *Science*, 2016](https://www.ncbi.nlm.nih.gov/pubmed/27034378))
(from the Supplementary Materials of [Nielsen et al, *Science*, 2016](https://www.ncbi.nlm.nih.gov/pubmed/27034378))








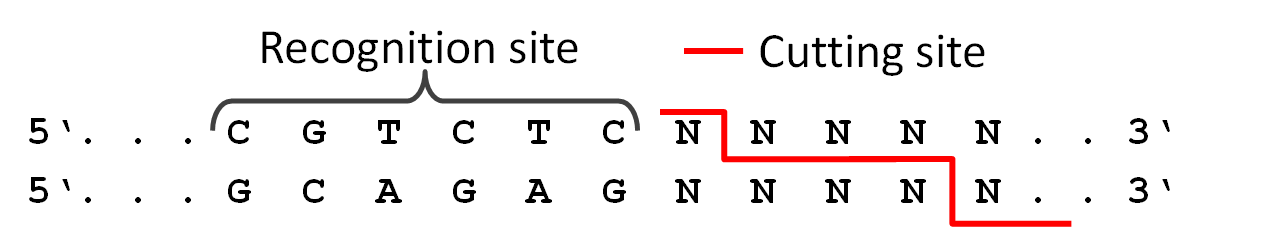

This SEP proposes a systematic set of "stem-top" glyphs representing small sites affecting DNA, RNA, or protein. The glyphs in this system are Biopolymer Location, Stability Element, and Cleavage Site.
Please see the full proposal at: https://github.com/SynBioDex/SBOLv-realizations/blob/develop/SEPs/SEP_V007.md