pinging @lsetiawan; remember that he's the real expert here on technical aspects of the JSON files and the climatology overlays. Don, please reply as needed.
confirmation if this is indeed the color ramp to be using for the climate rasters (both precip and temp)
Based on the "ppt" file prefix, http://data.nanoos.org/files/cz/mapoverlays/ppt_colorbar.png must be the color ramp for precipitation only. The colorbar image for each overlay is referenced in the overlay's JSON file .
Is it important or meaningful to use the same color breaks as the previous tiles? Or could we use a color scheme more consistent with the new app?
That's not entirely for me to answer. @aufdenkampe? But what I will say is that we spent a lot of time refining the colormaps and value ranges to work well for the variability found spatially and temporally, and based on common approaches for this type of data. And they've had some public exposure already, with generally positive feedback. For the MODIS EVI, I would strongly suggest that we stick with what we have. More broadly speaking, I would recommend using what we developed (color ramps and color breaks) and reassess in the future if needed and based on wider user input. That also would free you up at this time to focus on implementation challenges rather than rendering and styling decisions.
I'm also not quite sure what you mean by tuning color schemes of data to be more consistent with the app. With more overlay layers, in my experience the driving goals are instead distinctiveness across layers, coherence with common rendering practices in a core community, and a balanced representation of variability across the spatial and temporal domains.
appropriate raster values to assign to color breaks
I'm not sure I understand the question. Those values are defined in the JSON files. I think @lsetiawan explained the file a while ago, but if not, let us know and he can do that.


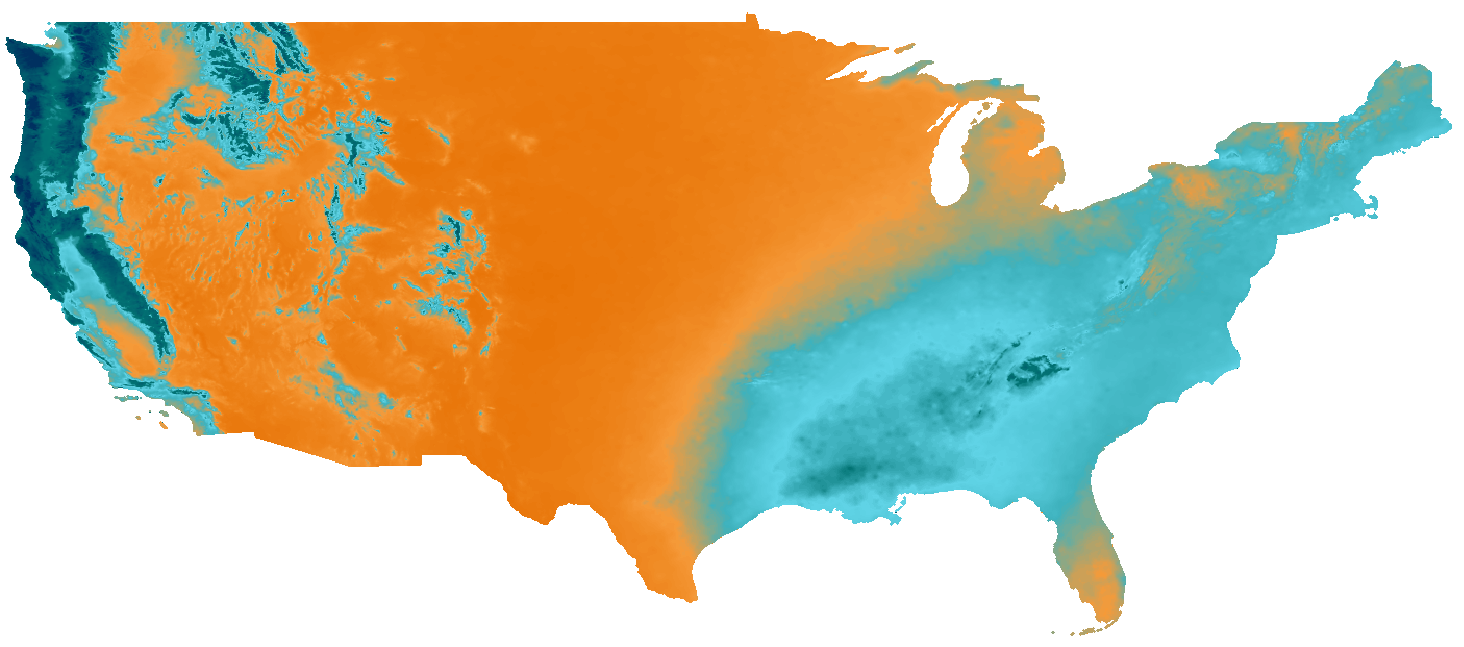

We are ingesting the BiG CZ climate rasters into GeoTrellis to create visual tiles. We need clarification from @emiliom as to the appropriate color ramp and color breaks for the rasters. There is some configuration in the sample JSON in the documentation that references this 1000×1px image: http://data.nanoos.org/files/cz/mapoverlays/ppt_colorbar.png
This could be the desired color ramp - we extracted 243 unique colors from the image to use. However, we need:
@emiliom Can you give us some guidance on the two questions above?