Sure thing! You can do that using the make_glcm and glcm_metrics functions. The shift parameter defines what pixels are considered neighbors.
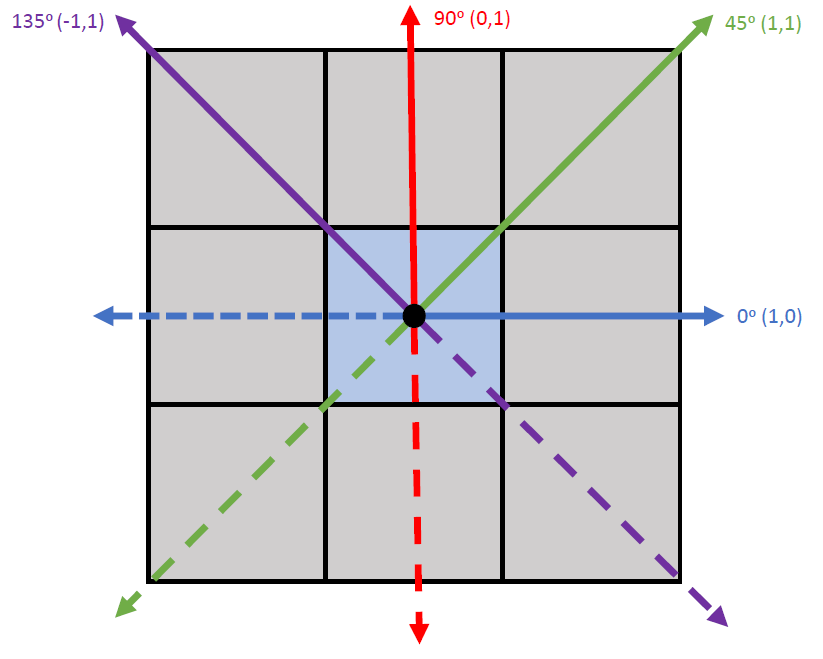
There isn't a "jump" parameter, but we can tabulate a GLCM for the entire image and then calculate the various measures from there. For example, below is code for a 200 x 200 matrix with 4 grey levels and a 0, 45, 90, and 135 degree shift. You can use these shifts individually or average them. Also, although typically neighboring pixels are defined as being one pixel away in a given direction you could change that to any number (e.g. two away horizontally would be shift = c(2,0)) You can also check out the section working with test_matrix in the ReadMe or look at the tutorial by Hall-Beyer.
library(GLCMTextures)
#> Loading required package: terra
#> Warning: package 'terra' was built under R version 4.2.2
#> terra 1.7.19
set.seed(5)
img<- matrix(data = sample(x = 0:3, size = 40000, replace = TRUE),
nrow = 200, ncol = 200) # 200 x 200 matrix with 4 grey levels
# Plot of image
plot(rast(img), col= grey.colors(100))
# Tabulate the GLCM for horizontal (zero degree shift)
GLCM_0<- make_glcm(img, n_levels = 4, shift = c(1,0), normalize = TRUE)
GLCM_0
#> [,1] [,2] [,3] [,4]
#> [1,] 0.06206030 0.06187186 0.06263819 0.06202261
#> [2,] 0.06187186 0.06168342 0.06305276 0.06233668
#> [3,] 0.06263819 0.06305276 0.06567839 0.06228643
#> [4,] 0.06202261 0.06233668 0.06228643 0.06216080
# Calculate the measures from the GLCM
results_0<- glcm_metrics(GLCM_0)
results_0
#> glcm_contrast glcm_dissimilarity glcm_homogeneity glcm_ASM
#> 2.4906281407 1.2464572864 0.5011884422 0.0625131161
#> glcm_entropy glcm_mean glcm_variance glcm_correlation
#> 2.7724850833 1.5026758794 1.2447918346 -0.0004195366
# Other shifts
GLCM_45<- make_glcm(img, n_levels = 4, shift = c(1,1), normalize = TRUE)
results_45<- glcm_metrics(GLCM_45)
GLCM_90<- make_glcm(img, n_levels = 4, shift = c(0,1), normalize = TRUE)
results_90<- glcm_metrics(GLCM_90)
GLCM_135<- make_glcm(img, n_levels = 4, shift = c(-1,1), normalize = TRUE)
results_135<- glcm_metrics(GLCM_135)
# Average across shifts
all_results<- rbind(results_0, results_45, results_90, results_135)
all_results
#> glcm_contrast glcm_dissimilarity glcm_homogeneity glcm_ASM
#> results_0 2.490628 1.246457 0.5011884 0.06251312
#> results_45 2.472387 1.242671 0.5016363 0.06252164
#> results_90 2.466307 1.238317 0.5036407 0.06252931
#> results_135 2.474306 1.240903 0.5028888 0.06252321
#> glcm_entropy glcm_mean glcm_variance glcm_correlation
#> results_0 2.772485 1.502676 1.244792 -0.0004195366
#> results_45 2.772416 1.502525 1.244640 0.0067864533
#> results_90 2.772355 1.502952 1.244640 0.0092285829
#> results_135 2.772403 1.502550 1.244615 0.0059952210
mean_results<- apply(all_results, MARGIN = 2, FUN = mean)
mean_results
#> glcm_contrast glcm_dissimilarity glcm_homogeneity glcm_ASM
#> 2.47590698 1.24208688 0.50233857 0.06252182
#> glcm_entropy glcm_mean glcm_variance glcm_correlation
#> 2.77241489 1.50267594 1.24467161 0.00539768Created on 2023-03-14 with reprex v2.0.2
I have a set of little images (200 x 200) of "uniform" textures and would like to calculate single GLCM sets of metrics for each image, instead of a raster with the values for each window position. Is it possible to set the parameters so that only one single GLCM is built for the entire image (thus only 1 set of metrics for the entire image) instead of having one GLCM (and one set of metrics) calculated for each window position?