Hi @maxentile, I see this issue and thank you so much for this very detailed explanation. Your feedback is important to us. I will try to reproduce and get back to you as soon as possible.
Closed maxentile closed 5 years ago
Hi @maxentile, I see this issue and thank you so much for this very detailed explanation. Your feedback is important to us. I will try to reproduce and get back to you as soon as possible.
Hi @maxentile, I don't quite understand the difference on how you get the figure in 1.1 and in 2.1. I tried to reproduce using the code below:
import torchani
import torch
import math
%pylab inline
device = torch.device('cpu')
model = torchani.models.ANI1ccx()
species = model.species_to_tensor('CHHHH').to(device).unsqueeze(0)
coordinates = torch.tensor(
[[ 0. , 0. , 0. ],
[ 0.63918859, 0.63918859, 0.63918859],
[-0.63918859, -0.63918859, 0.63918859],
[-0.63918859, 0.63918859, -0.63918859],
[ 0.63918859, -0.63918859, -0.63918859]])
global_lowest_energy = -106313.8125
def try_range(start, end):
scale = torch.arange(start, end, 0.005)
bond_lengths = 0.63918859 * math.sqrt(3) * scale
species_ = species.expand(scale.shape[0], -1)
coordinates_ = coordinates * scale.unsqueeze(-1).unsqueeze(-1)
_, energies = model((species_, coordinates_))
energies = energies * 2625.50 - global_lowest_energy # Hartree to kJ/mol
plt.plot(bond_lengths.numpy(), energies.detach().numpy())
plt.title('ANI-1ccx')
plt.xlabel('C-H length (Angstrom)')
plt.ylabel('Energy (kJ/mol)')
plt.show()
try_range(0, 5)
try_range(0.8, 1.2)and I get


which is similar to your figure at 2.1.
But how do you get the figure at 1.1?
Thanks a lot!
Dear Xiang, thank you very much for looking into this! Apologies for my delayed response.
Here is the script I used to reproduce the energy scans: https://gist.github.com/maxentile/01eb9c09ad6265eaa316efc4078a3cc7
and the output:

Hey @maxentile @wiederm - first of all, thanks for very detailed "post mortem". We are investigating... Few comments.
Current "pure" ML potentials have no physically imposed constraints. Therefore, one has to use them exactly within their domains of applicability. For ANI at this point, it's just a single molecule potential. We are working to include bond breaking, but the current public model has sampled C-H bonds within [~0.8... ~1.6] with closed shell hamiltonian. As you see it produces a realistic potential well. We are aware of artificial short distance minima - it will be fixed soon. However, to describe the proper bond breaking reactions requires sampling with unrestricted open shell Hamiltonian, proper spin states, etc.
The transfer learning model is bleeding edge research, and we are still looking for some anomalies because it's essentially a Frankenstein model between DFT and CCSDT. Did you try pure DFT model (1x)?
By the nature ML potentials are reactive, so you can't just use a 1-2fs step for MD. There is also no SHAKE. Realistic time steps should be like in AIMD: 0.1...0.4 fs depending on the fastest vibration in the system and temperature.
Thanks for the detailed response, @isayev!
We're just trying to compute the free energy between two tautomers at this point, which should involve only two well-defined end-states that should be fully within the domain of applicability of the model: We're essentially integrating the partition function over the thermally-accessible conformations of the fully closed-shell Hamiltonian. The method we're using to do so may break bonds or alchemically annihilate atoms, but since we're doing this with classical statistical mechanics and free energy is a state function, all we need is well-behaved chemical end-states.
Because high-quality Langevin integrators like VRORV (BAOAB) integrators can be pushed to their stability limit without creating large errors in sampling from conformational probability densities, and our goal is only to compute relative free energies in configuration space, I don't think we're bound by the need to use tiny timesteps as in AIMD. That assumes there are no nightmarish pathologies introduced into the energy landscape---those energy cusps worry me---that would cause the superconvergence properties to break down.
@jchodera yah, those look pretty bad! Could you please try "normal" DFT trained ANI? Also, irrespective of integrators you really need a shorter timer step for those potentials. I have not tried VRORV, but we use 0.5fs time step for MD.
Hi all,
Thank you for your interest in our work, and for sharing your experience with us. I'd like to throw in my 2-cents. This post freaked me out a bit so I might have gone a little overboard attempting validate @maxentile results. My findings in short: NeuroChem/ASE with ANI-1ccx are unable to reproduce @maxentile's results. This likely means that ANI-1ccx is not to blame, but some code implementation problem elsewhere. Keep reading for details.
I would like to clarify a few things with regard to how ML potentials work. It should not surprise anybody that an ML potential can have a global minimum far lower in energy than what is considered the physical minimum of a system. In fact, there are likely many such minima in the space of all possible input coordinates. Does it surprise me if there is a weird minima for methane that exists for ANI-1ccx? The answer is no. I can show an ML model meant to categorize pictures of horses a picture that to you and I appears to be static noise, but the model will categorize it as a horse. This is an inherent problem in all deep learning. What does surprise me is the 300K thermal accessibility of such states. Ensuring that an ML potential is applicable in MD simulation is a matter of sampling the physically relevant space of interest well enough that the probability of reaching non-physical states under the desired conditions (e.g. 300K temperature) approaches zero. Of course, this is assuming that you start from a semi-realistic geometry. As Olexandr pointed out, most of the regions of the methane potential surface discussed in this thread are well outside of any sampled space, as can be seen by the large standard deviation in @maxentile's figures. Fun fact, Gaussian 09 will crash if you were to give it a bond distance less than 0.5A, so we couldn't have sampled that if we wanted to. In the end, you should use the tools at your disposal, such as the ensemble disagreement to notify you when you do enter into these unphysical regions. With that said, I find it hard to believe (based on my intimate knowledge of the ANI data sets) methane in vacuum is a problem for ANI-1ccx, so I ran a few tests.
I wanted to validate whether the ability for @maxentile's MD to escape the well described region of chemical space was ANI-1ccx potential related, TorchANI related, or other code related. Therefore, I decided to run similar studies of methane independently, using our other code package (NeuroChem+ASE).
To see if I could reproduce the "bent" geometry energy minimum, or at least a minima, I copied @maxentile "bent" coordinates and ran a geometry optimization with ANI-1ccx in the NeuroChem/ASE package using LBFGS. Below is the output, where I print all C-H distances and H-C-H angles before and after the optimization:
Distances: [1.27001867 1.27055506 1.06795258 1.06973148] Angles: [ 92.17736159 102.57026975 102.53879659 102.57196359 102.52411243 143.47920595] LBFGS: 1 15:11:31 -1100.161277 6.5967 LBFGS: 2 15:11:31 -1100.512521 5.2980 LBFGS: 3 15:11:31 -1100.790695 3.4940 LBFGS: 4 15:11:31 -1100.966503 1.3452 LBFGS: 5 15:11:31 -1101.040357 0.7483 LBFGS: 6 15:11:31 -1101.062402 0.6299 LBFGS: 7 15:11:31 -1101.091741 0.2075 LBFGS: 8 15:11:31 -1101.093479 0.1288 LBFGS: 9 15:11:31 -1101.094436 0.0762 LBFGS: 10 15:11:31 -1101.094680 0.0649 LBFGS: 11 15:11:31 -1101.095067 0.0416 LBFGS: 12 15:11:31 -1101.095151 0.0197 LBFGS: 13 15:11:31 -1101.095163 0.0024 LBFGS: 14 15:11:31 -1101.095164 0.0001 [ANI Total time: 0.09245634078979492 seconds] Distances: [1.08765633 1.08765634 1.08766003 1.08765931] Angles: [109.46917795 109.47189449 109.47154575 109.47178379 109.47151341 109.47164326]
As you can see, the geometry optimization returned the "bent" structure to the correct tetrahedral geometry. I then did a similar stretch of the tetrahedral vs bent geometry (Section 1.4) and I am not seeing the same behavior. See the figure below:

Here the bent geometry is higher in energy than the tetrahedral geometry, as we would expect. I have also not seen any of the strange cusps that @jchodera pointed out.
The first thing I noticed was that my ANI-1ccx geometry optimization resulted in a minimum energy bond distance of 1.087A for the tetrahedral geometry, which seems to be different from what @maxentile found with his scan. This difference needs to be investigated. Below I provide a tetrahedral stretch similar to what @maxentile provided. This clearly shows a lower bond distance than what @maxentile presented: 1.107A.

What interests me more than the existence of lower energy minima is the accessibility of those minima. Therefore, I carried out an MD study similar to @maxentile using the Langevin thermostat as implemented in ASE. I stuck with a 1fs time step as @maxentile did, even though I would recommend smaller since we have ran into trouble in the past mixing > 0.5fs time steps and hydrogens. I tried running longer timescale simulations than @maxentile, more specifically 250ps at 300K but I could not reproduce the issue of finding a bent state, or anything other than the tetrahedral geometry. I then tried 250ps at 400K, then 500K, and I still could not reproduce this behavior. I have ran the simulation around 4 times at each temperature using different random seeds. I was have not been able to reproduced it. Below are similar figures to what @maxentile originally provided for his MD. The black dashed lines represent the optimized geometry values.
C-H bond distance along the trajectory:

C-H bond distance histogram (data from the entire trajectory):

Here we can see that the bond distance correctly fluctuates around the equilibrium bond distance. It is slightly off center likely due to the anharmonicity of the bond.
H-C-H angle along the trajectory:

H-C-H angle histogram (data from the entire trajectory):

The angle correctly fluctuates around the equilibrium angle.
Energy along the trajectory:

The energy of the trajectory never drops below the optimized tetrahedral geometry energy.
Given I cannot reproduce this bent structure in MD or optimization with ANI-1ccx and NeuroChem/ASE points to possible implementation problems with either the thermostat or TorchANI. I believe the above evidence shows this is not a problem with the potential itself. Also, we have validated that TorchANI and NeuroChem agree to single precision, but we should still test TorchANI to be safe. @zasdfgbnm I would recommend getting someone to run a similar study as I did above with TorchANI/ASE to make sure that we agree. If we do agree then there must be something specific to @maxentile code causing the problem.
It is possible that methods for accelerating the discovery of low energy states might find their way out of a well described physical minima. In the event that happens, you can likely tell by a large ensemble disagreement. At that point the fix would be to rerun active learning using the accelerated sampler to better sample conformational space.
Dear all,
I'm extremely extremely sorry for this!
When searching for the source of the discrepancy between @Jussmith01 's and my results, I have found the source of the discrepancy. I had made a change to the aev.py file in my local installation weeks ago and had not reverted it, causing a severe bug in how the atomic environment vectors were computed, invalidating everything in point 1 of my original issue. (Plots in point 2 were produced by @wiederm and were not affected by this, but you mention you are already aware of and addressing the issue of short-distance artificial minima.)
With a fresh installation, this test
import torchani
import torch
import numpy as np
device = torch.device('cpu')
# ANI-1x model
model_ANI1x = torchani.models.ANI1x()
species_ANI1x = model_ANI1x.species_to_tensor('CHHHH').to(device).unsqueeze(0)
# ANI-1ccx model
model_ANI1ccx = torchani.models.ANI1ccx()
species_ANI1ccx = model_ANI1ccx.species_to_tensor('CHHHH').to(device).unsqueeze(0)
def ANI1x_energy(x_in_angstroms):
coordinates_in_angstroms = torch.tensor([x_in_angstroms],
requires_grad=False, device=device, dtype=torch.float32)
_, energy_in_hartree = model_ANI1x((species_ANI1x, coordinates_in_angstroms))
return float(energy_in_hartree.detach())
def ANI1ccx_energy(x_in_angstroms):
"""Remove units code"""
coordinates_in_angstroms = torch.tensor([x_in_angstroms],
requires_grad=False, device=device, dtype=torch.float32)
_, energy_in_hartree = model_ANI1ccx((species_ANI1ccx, coordinates_in_angstroms))
return float(energy_in_hartree.detach())
# tetrahedron with bond lengths 1.08765633 A
x_tetrahedral = np.array([[ 0. , 0. , 0. ],
[ 0.62795867, 0.62795867, 0.62795867],
[-0.62795867, -0.62795867, 0.62795867],
[-0.62795867, 0.62795867, -0.62795867],
[ 0.62795867, -0.62795867, -0.62795867]])
x_bent = np.array([[ 0. , 0. , 0. ],
[ 0.64702535, 0.64647233, 0.8811238 ],
[-0.64734113, -0.6475463 , 0.8808765 ],
[-0.7168874 , 0.7172707 , -0.33484012],
[ 0.7182246 , -0.71852016, -0.33497408]])
hartree_to_kJ_mol = 2625.499638
print('ANI1x(tetrahedral) - ANI1x(bent): {:.3f} kJ/mol'.format((ANI1x_energy(x_tetrahedral) - ANI1x_energy(x_bent))* hartree_to_kJ_mol))
print('ANI1ccx(tetrahedral) - ANI1ccx(bent): {:.3f} kJ/mol'.format((ANI1ccx_energy(x_tetrahedral) - ANI1ccx_energy(x_bent)) * hartree_to_kJ_mol))outputs
ANI1x(tetrahedral) - ANI1x(bent): -134.628 kJ/mol
ANI1ccx(tetrahedral) - ANI1ccx(bent): -129.350 kJ/molwhere before, on my buggy local installation, it was showing that the bent geometry was more favorable.
Again, I am extremely sorry for this!!! This was a mistake in my local installation, not in torchani.
I am closing this issue while I investigate further.
Thank you very much to @zasdfgbnm @Jussmith01 @isayev for all your quick and insightful feedback, especially on a weekend, and I sincerely apologize for having freaked you out about an issue that was 100% my mistake!!
Hello,
Thank you very much for making this project available!
We are excited about the prospect of using ANI-1ccx for tautomer ratio prediction, but have started encountering some surprising behavior when designing and testing Monte Carlo moves for this purpose.
Tagging Marcus Wieder (@wiederm) who helped prepare this Issue with me, and John Chodera (@jchodera) who may be interested in following this thread.
1. Existence and favorability of unphysical local minima on methane example
1.1. Bond-length scan from a tetrahedral geometry
We started by inspecting the methane example
If we take an ideal tetrahedral geometry and scale all the C-H bond lengths, we obtain the following energy profile:
The minimum-energy tetrahedral geometry of methane along this scan is approximately:
with a C-H bond length of 1.107 angstroms, and an energy of -106266.1875 kJ/mol.
1.2. Accessibility of unphysical local minima in room-temperature MD
We ran 50 picoseconds of constant-temperature MD at 300K starting from the tetrahedral geometry obtained above.
(Code: https://gist.github.com/maxentile/ac04d1fab8dd4aa217fe224190d06765)
The simulation quickly finds many geometries with lower apparent energy than the starting tetrahedral geometry.
The simulation does not retain a tetrahedral geometry. Instead the H-C-H angles appear to be metastable.
Here are the histograms of H-C-H angles visited in the trajectory:
Also the C-H distances appear to be bistable: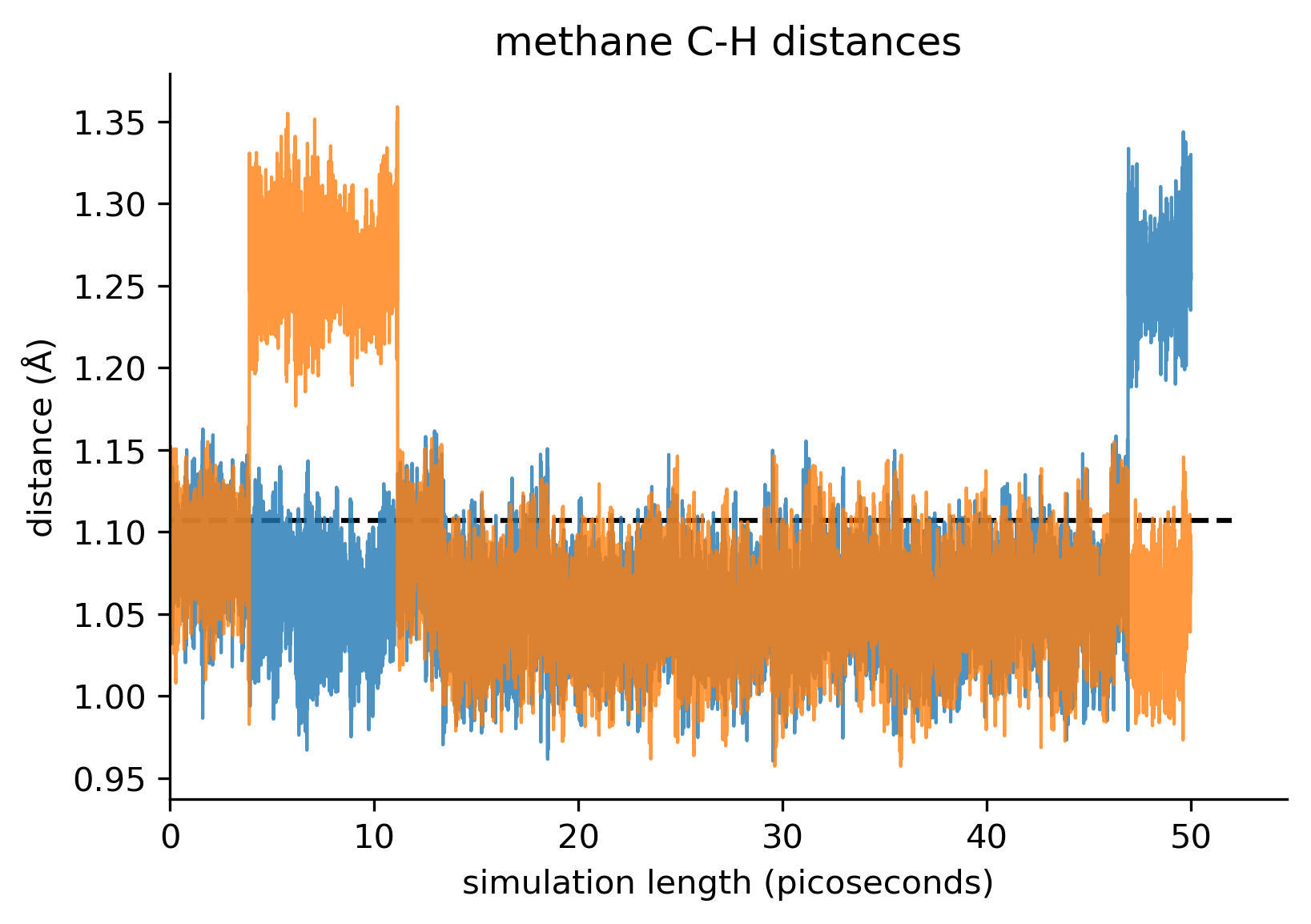
1.3. ANI-1ccx predicts a non-tetrahedral geometry to have nearly 50 kJ/mol lower energy than the minimum-energy tetrahedral geometry
The lowest-energy snapshot from this trajectory includes nearly right angles, and is over 40 kJ/mol more favorable than the minimum-energy tetrahedral geometry. We further minimized its energy using gradient descent on the ANI-1ccx potential, obtaining the following:
with two of the C-H bonds at length 1.27 angstroms and the other two C-H bonds at length 1.07 angstroms.
The H-C-H angles at this conformation are 92.2 , 102.5, and 143.5 degrees.
The ANI-1ccx energy of this configuration is -106313.8125 kJ/mol.
This is ~47 kJ/mol more favorable than the minimum-energy tetrahedral geometry.
Is there an intuition for this behavior, and is anything similar observed for more complicated molecules?
1.4. Energy scans
We took the energy-minimized geometry and scaled it up and down, obtaining the following energy profile (in orange):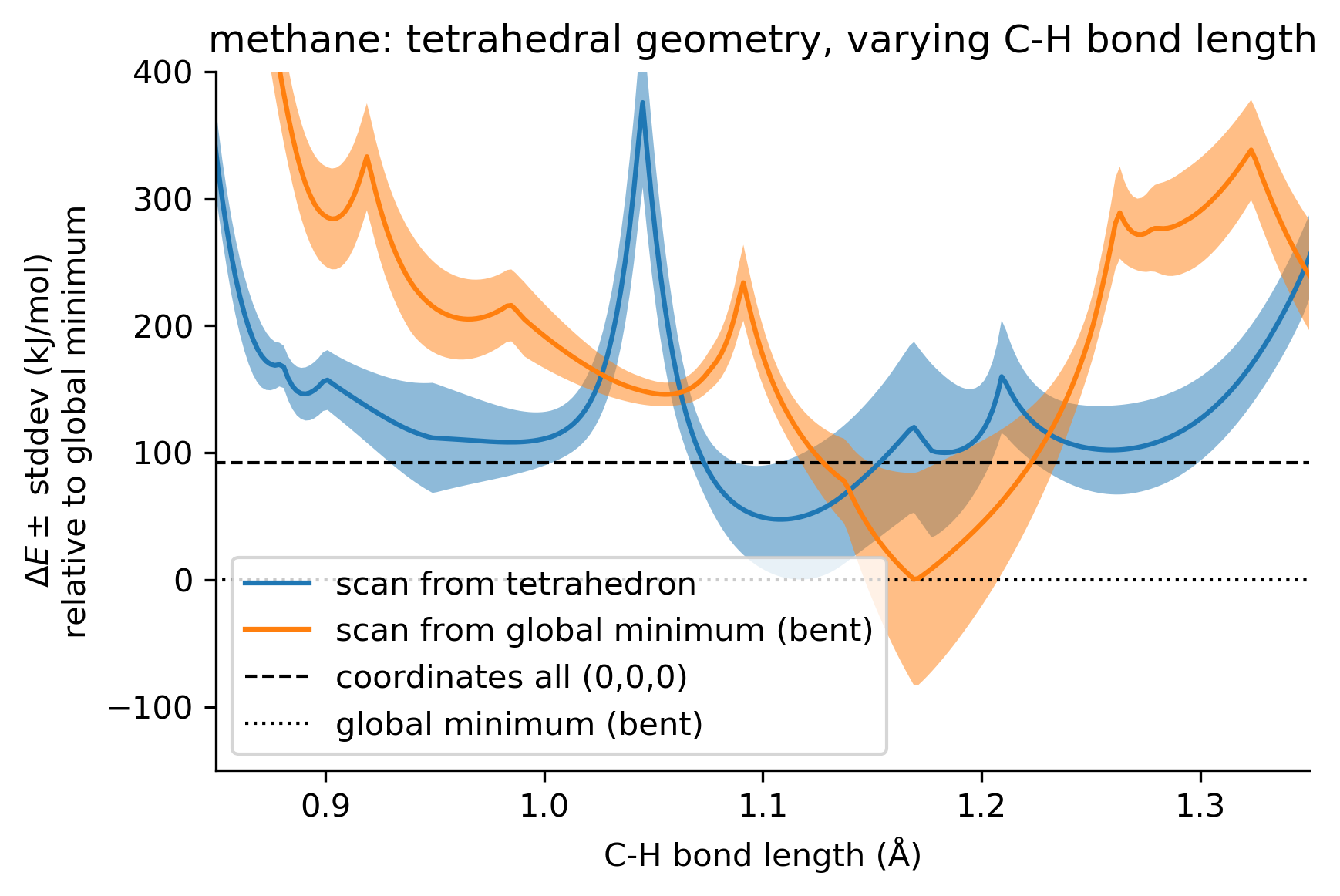
The x-axis represents the average C-H bond length. The uncertainty bands represent the standard deviation among the 8 models in the ANI-1ccx ensemble.
For comparison, in blue is the energy profile we obtained for tetrahedral geometries, the dashed line is the energy obtained when the coordinates are all (0,0,0), and the dotted line at 0 corresponds to the global minimum energy encountered so far.
We observe that the energy profiles have many local minima and cusps. Is there an intuition for why the energy profile looks this way, or how it might be corrected?
Also, it appears that the ANI-1ccx ensemble has large uncertainty throughout these scans -- is there guidance on how to use this information?
2. Local energy minimum when all coordinates are zero
We further noticed that for the methane example in the
Computing Energy and Forcetutorial https://aiqm.github.io/torchani/examples/energy_force.html, setting all coordinates to (0,0,0) is ~30kJ/mol more favorable than the the starting geometry (-106221.367 kJ/mol vs -106189.195 kJ/mol).2.1. Energy scan, scaling all C-H distances
For illustration, here is the energy profile as we scale the example starting geometry up and down: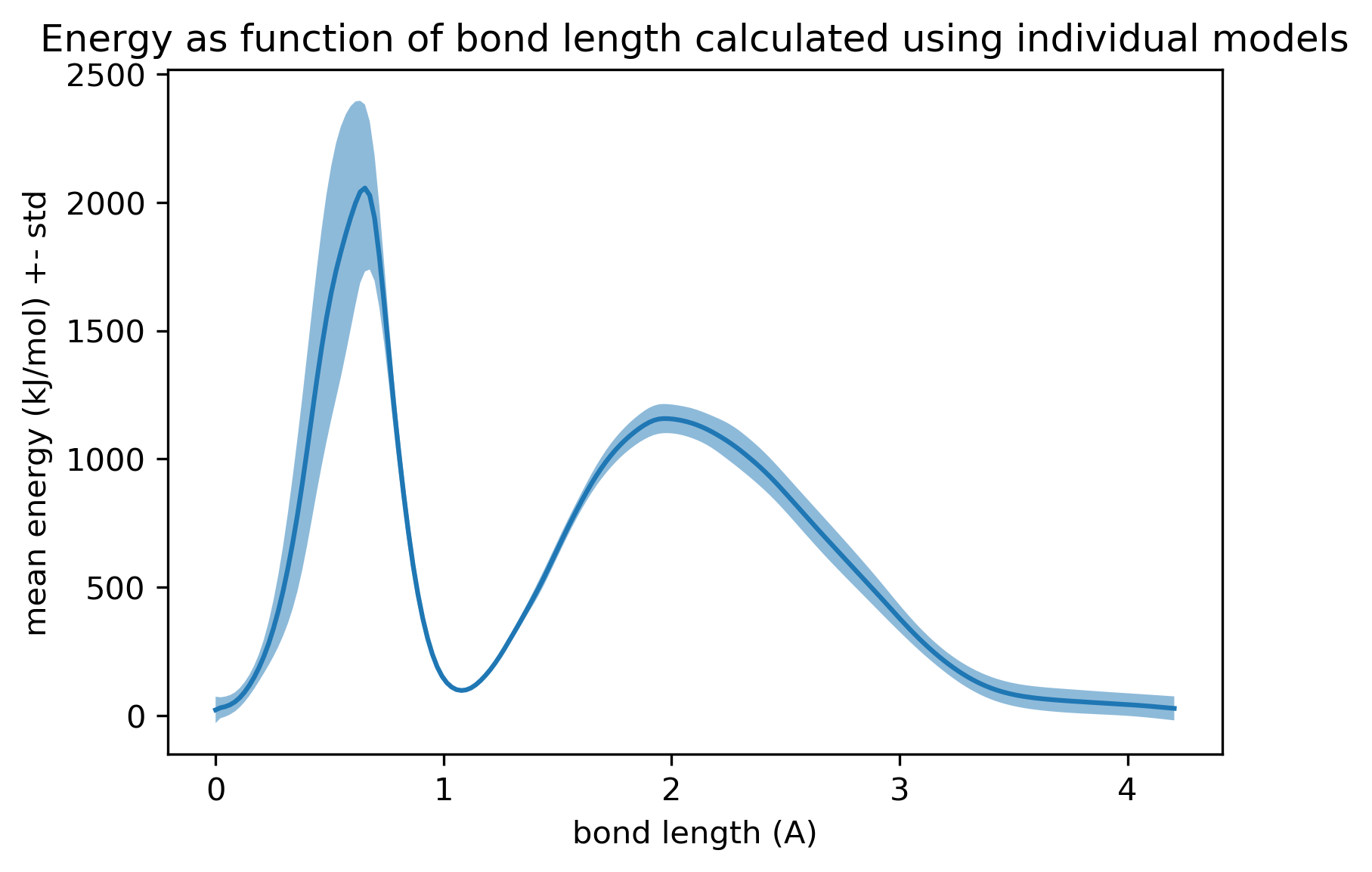
The existence of a local minimum with all nuclei on top of each other is concerning in the context of Monte Carlo moves that instantaneously propose to move hydrogens, since they might randomly propose geometries that induce steric clashes, and we would want such proposals to be rejected.
2.2. Comparison: changing only one H position, rather than all H positions
If we bring only one of the hydrogens closer to the central carbon (rather than all of them), we obtain an energy profile that instead looks like this: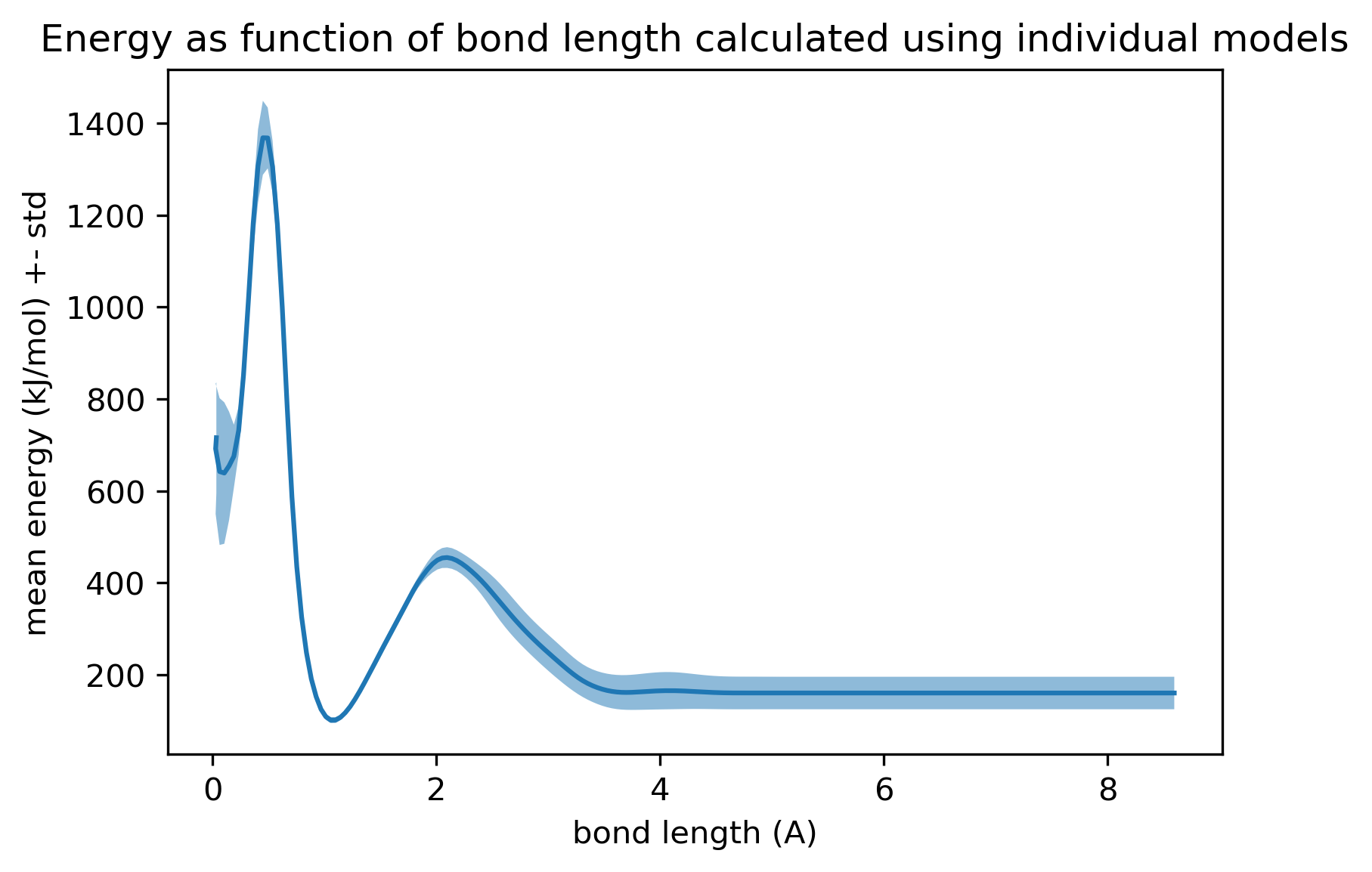
The high energy predicted when one hydrogen is placed on top of the central carbon is reassuring, since it suggests instantaneous Monte Carlo proposals that try to place one hydrogen directly on top of a heavy atom will not be accepted with any appreciable probability. However, it is still surprising that the energy does not increase monotonically as the interatomic distance approaches zero from the equilibrium bond length.
Is there an intuition for how to correct this behavior?
Best regards, Josh