Hi, there are a few things going on here. First, the only reason the data are rescaled is for visualization purposes, ie. to plot all the sites against a common set of centiles (otherwise each site will be different). If you plot each site separately it is not necessary. Also you probably do not even need to worry about site effects if you are using UKB data and a phenotype like that.
Second, it is important to understand that the warp implicitly uses a SHASH distribution which is good for continuous data and can model many shapes. But there are also a lot of distributions it cannot fit and for the interval data you show (arcuate) you will never achieve an acceptable fit. You would be better to use a beta distribution instead which is not currently supported in pcntoolkit but might be at some point. See here (and the references therein) for more details about the SHASH dist: https://www.biorxiv.org/content/10.1101/2022.10.05.510988v2
The last example is an optimization failure which you might be able to fix by changing the optimizer or regularization settings. You also should know that fitting shape parameters is a very hard optimization problem in general because of strong dependencies between parameters and often weak identifiability of different parameter values.
Hi again,
I am facing a follow-up problem to a previous issue (https://github.com/amarquand/PCNtoolkit/issues/114) with regard to the warping process. As noted there, I have amended my code as recommended by adding "warp='WarpSinArcsinh', warp_reparam= True" to the estimate function to perform warped BLR.
My aim with this analysis is to get centile curves of WMH and related markers as well as respective z-scores for downstream predictive modelling in a sample from two cohorts.
Currently I am trying to reproduce the centile curves for WMH volume with warped BLR as presented in this manuscript (https://www.sciencedirect.com/science/article/pii/S1053811921009873#fig0004) as it accounts for potential non-gaussianity in the data. Therefore, I am following this tutorial "https://github.com/predictive-clinical-neuroscience/PCNtoolkit-demo/blob/main/tutorials/BLR_protocol/transfer_pretrained_normative_models.ipynb". The tutorials mentions that the predictions (yhat, S2) from the dummy model are after the prediction in the warped space and need to be inversely warped to achieve plotting in input space. Apparently, also the true data (y_te) is rescaled in the tutorial.
Now I am wondering whether rescaling the true data is necessary for my usecase. I have noticed that applying this code, the resulting data are shifted in the y direction compared to a plot of the raw data for some of the imaging-derived phenotypes I am investigating. Furthermore, the centile curves do not look as I would expect.
Here the plot resulting from the abovementioned code for plotting the WMH volume which appears as expected.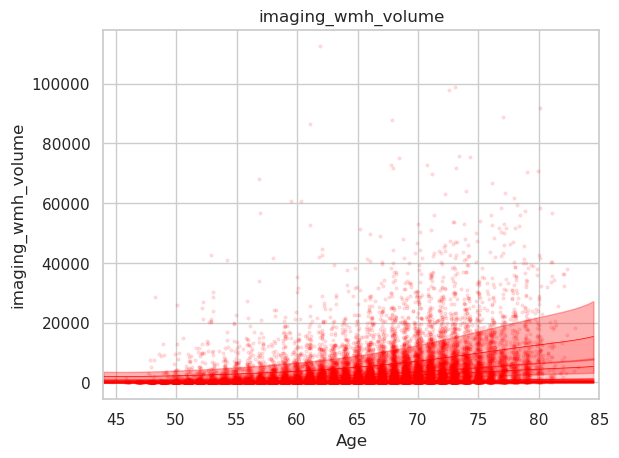
Here a simple scatterplot of the raw data.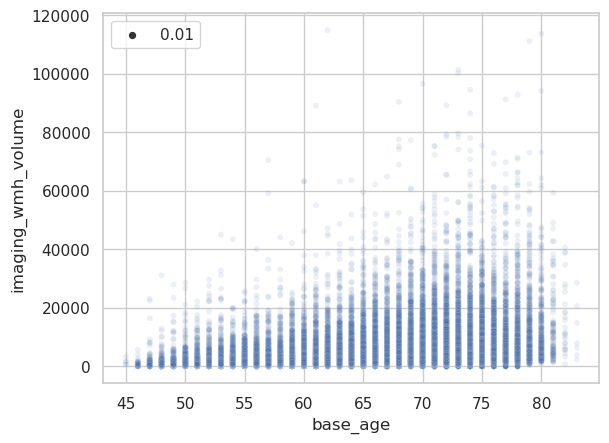
The problem is apparent when looking at other imaging-derived phenotypes.
Disconnectivity of the arcuate fascicle in percent using the abovementioned plotting code. Note that the data is shifted upwards (y interval is 20-120% instead of 0-100%).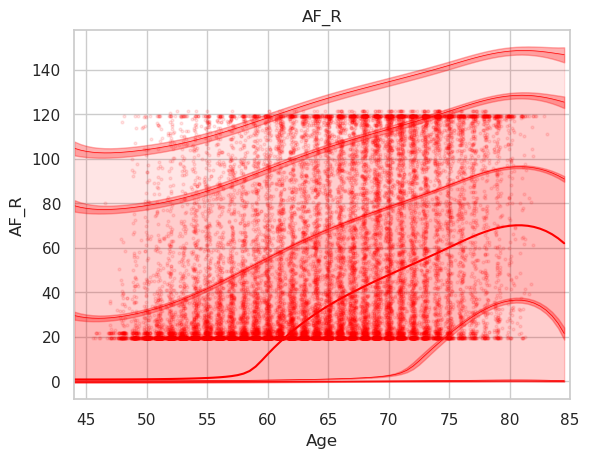
And the corresponding raw data plot.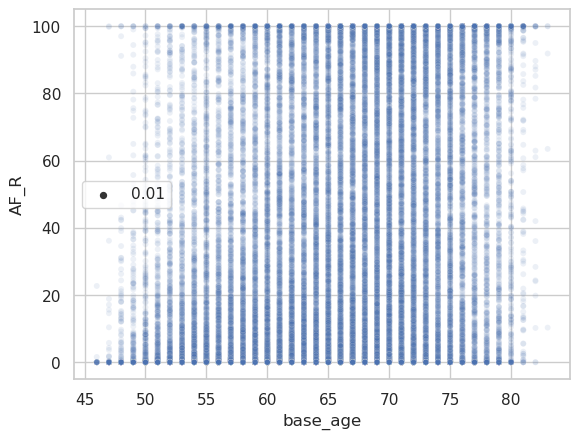
The same plots for the peak width of skeletonized mean diffusivity (PSMD).
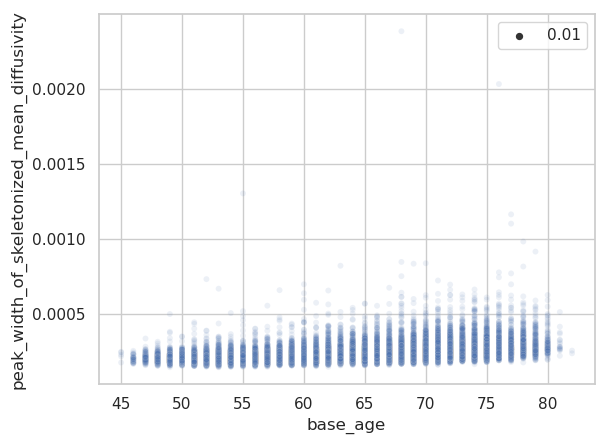
Are there some assumptions that need to be met to apply warped BLR when modelling a variable, like is non-gaussianity of residuals required? One difference to the tutorials is that I use a 2-fold cross-validation via
estimate()to get zscores for all individuals. I noticed that the resulting centile curves differ relevantly if I rerun the analysis. Maybe because of probabilistic sampling of training and test set during CV?The complete code I use can be found in this jupyter notebook: https://drive.google.com/file/d/1p0jHzDC832yVKWd7p0PULgfhbnnYUi0F/view?usp=sharing.
I would be very grateful to get some help. Happy to provide further information if required.
Thanks a lot in advance!
Marvin