ASF subversion and git services (migrated from JIRA)
Commit 2aba8b89796150760fa8939452b5d9271d7b5b7b in lucene-solr's branch refs/heads/LUCENE-9004 from Michael Sokolov https://gitbox.apache.org/repos/asf?p=lucene-solr.git;h=2aba8b8
LUCENE-9004: approximate nearest vector search (WIP)
"Semantic" search based on machine-learned vector "embeddings" representing terms, queries and documents is becoming a must-have feature for a modern search engine. SOLR-12890 is exploring various approaches to this, including providing vector-based scoring functions. This is a spinoff issue from that.
The idea here is to explore approximate nearest-neighbor search. Researchers have found an approach based on navigating a graph that partially encodes the nearest neighbor relation at multiple scales can provide accuracy > 95% (as compared to exact nearest neighbor calculations) at a reasonable cost. This issue will explore implementing HNSW (hierarchical navigable small-world) graphs for the purpose of approximate nearest vector search (often referred to as KNN or k-nearest-neighbor search).
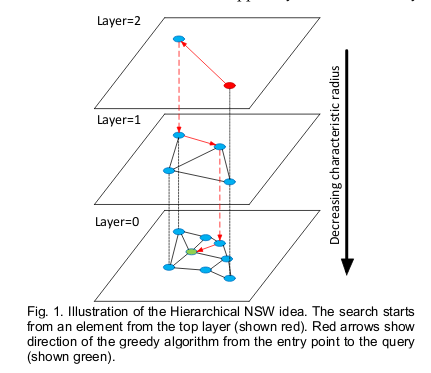
At a high level the way this algorithm works is this. First assume you have a graph that has a partial encoding of the nearest neighbor relation, with some short and some long-distance links. If this graph is built in the right way (has the hierarchical navigable small world property), then you can efficiently traverse it to find nearest neighbors (approximately) in log N time where N is the number of nodes in the graph. I believe this idea was pioneered in [1]. The great insight in that paper is that if you use the graph search algorithm to find the K nearest neighbors of a new document while indexing, and then link those neighbors (undirectedly, ie both ways) to the new document, then the graph that emerges will have the desired properties.
The implementation I propose for Lucene is as follows. We need two new data structures to encode the vectors and the graph. We can encode vectors using a light wrapper around
BinaryDocValues(we also want to encode the vector dimension and have efficient conversion from bytes to floats). For the graph we can useSortedNumericDocValueswhere the values we encode are the docids of the related documents. Encoding the interdocument relations using docids directly will make it relatively fast to traverse the graph since we won't need to lookup through an id-field indirection. This choice limits us to building a graph-per-segment since it would be impractical to maintain a global graph for the whole index in the face of segment merges. However graph-per-segment is a very natural at search time - we can traverse each segments' graph independently and merge results as we do today for term-based search.At index time, however, merging graphs is somewhat challenging. While indexing we build a graph incrementally, performing searches to construct links among neighbors. When merging segments we must construct a new graph containing elements of all the merged segments. Ideally we would somehow preserve the work done when building the initial graphs, but at least as a start I'd propose we construct a new graph from scratch when merging. The process is going to be limited, at least initially, to graphs that can fit in RAM since we require random access to the entire graph while constructing it: In order to add links bidirectionally we must continually update existing documents.
I think we want to express this API to users as a single joint
KnnGraphFieldabstraction that joins together the vectors and the graph as a single joint field type. Mostly it just looks like a vector-valued field, but has this graph attached to it.I'll push a branch with my POC and would love to hear comments. It has many nocommits, basic design is not really set, there is no Query implementation and no integration iwth IndexSearcher, but it does work by some measure using a standalone test class. I've tested with uniform random vectors and on my laptop indexed 10K documents in around 10 seconds and searched them at 95% recall (compared with exact nearest-neighbor baseline) at around 250 QPS. I haven't made any attempt to use multithreaded search for this, but it is amenable to per-segment concurrency.
[1] https://www.semanticscholar.org/paper/Efficient-and-robust-approximate-nearest-neighbor-Malkov-Yashunin/699a2e3b653c69aff5cf7a9923793b974f8ca164
UPDATES:
Migrated from LUCENE-9004 by Michael Sokolov (@msokolov), 16 votes, resolved Dec 30 2020 Attachments: hnsw_layered_graph.png Linked issues: