Hi @HectorM14 thanks for this contribution, the plot looks really nice. It will be really great if you send a PR with this new plot. Please use our existing plots as a reference, maybe pair_plot is a good place to check.
Open hectormz opened 5 years ago
Hi @HectorM14 thanks for this contribution, the plot looks really nice. It will be really great if you send a PR with this new plot. Please use our existing plots as a reference, maybe pair_plot is a good place to check.
@aloctavodia I'll work on a PR that follows the current format as existing plots. Thanks! I'll check in if I have questions.
I volunteer to help with the PR. In fact this issue has given me the idea of extending plot pair using customizable functions.
There would be 3 input functions, each with its own kwargs:
The triangular functions should have as arguments values1, values2, ax, and the diagonal one values1, ax. If one of these functions were None, ax.axis("off") should be called.
The skeleton would be quite similar to plot_pair. Therefore, using plot_posterior as diagonal function and as lower triangular a wrapper on plot posterior similar to this:
def difference_hist(values1,values2,ax,**kwargs):
return az.plot_posterior(values1-values2,ax=ax,**kwargs)should generate your first output. Adding plot_kde or a wrapper on plot_pair as upper triangular function should generate your second output. The default could be a plot pair with the marginal distributions on the diagonal, and nothing on the upper triangle which is a quite common plot.
Hi @OriolAbril , I agree, this could be very clean and simple since it mostly relies on existing plotting functions in arviZ.
In my first draft of my function that I included, I was only interested in plotting kde/histograms, and I wanted to make all the arguments explicit for the user (me), instead of capturing everything as **kwargs and passing them to plot_posterior. Adding another triangle (scatter/kde/hexbin) because if all the passed arguments are explicit, the list grows a bit, OR we're capturing kwargs for the two triangles, and have to separate them before passing them along.
What do you think about that? Should there be many explicit and obvious options for the user, or should they know which arguments to pass?:
def plot_compare_posterior(data, var_name, lower='hist', upper='kde', **kwargs):As I previously thought about it, (compared to plot_posterior) a user would only want to compare a single var_name, and optionally provided a list of indices if they don't want to compare every item in the vectorized posterior.
What would be an appropriate name for this plot?
About the arguments I would start with:
def plot_pair_extended(
# arguments needed to mimic plot_pair
data, var_names=None, coords=None, figsize=None, ax=None,
# function arguments
lower_fun=plot_kde, upper_fun=None, diag_fun=plot_posterior,
# function kwargs, as the functions are independent, their kwargs should be too
lower_kwargs=None, upper_kwargs=None, diag_kwargs=None):The coords will allow to select some of the items in var_names. In addition however, it may be useful to compare different chains instead of all draws.
As for the name, I could not think of anything else other than pair_plot_extended. But I would not worry about it too much about it yet.
I was thinking that changing the arguments of the triangle functions to values, ax like in the diagonal case, with the difference of values being 2D instead of 1D will probably ease the default values and creating (or even better avoid creating) wrappers. For instance, plot_pair itself could be used as lower triangular function default. This would allow more freedom in the kind of pairplot used.
If I follow you correctly:
plot_posterior (if shown), and the user can still pass along specific kwargsplot_posterior (if shown), and the user can still pass along specific kwargsplot_pair (if shown), and the user can still pass along specific kwargsI think that is along the lines of what I was previously thinking. With that reasoning though, if I only wanted one of the triangles, the type of plot I want, and where I want it, may differ from someone else. But I think we could include some default boolean argument that handles that desire.
Referring to the triangle options by their position (upper/lower) is intuitive, but we could also refer to them as the 1D or 2D comparisons as you mentioned.
I did most of my work only in PyMC3 until recently, but I'm now seeing how great InferenceData and coords in arviZ are for these plots
Will coords allow the user to select particular (or all) chains to compare, or that is another matter that you were bringing up?
I had in mind to use functions as parameters, therefore, the diagonal and both triangular plotting functions can be any function defined by the user. That is why in the definition of plot_pair_extended nor plot_kde nor plot_posterior are strings. Therefore, a typical use case would be:
import arviz as az
# define simple function to plot the difference
def difference_hist(values1, values2, ax):
ax = az.plot_posterior(values1-values2, ax=ax, kind='hist')
return ax
data = az.load_arviz_data('centered_eight')
# call plot_pair extended overriding the default lower triangular function
az.plot_pair_extended(data, var_names=["mu"], lower_fun=difference_hist)There are no quotes around difference_hist in order to pass the function to plot_pair_extended Hence, calling lower_fun(x, y, ax) inside plot_pair_extended will be equivalent to calling difference_hist(x, y, ax) in the main program.
In order to leave one of the triangles or the diagonal empty, None could be passed as function (like the default for upper_fun) to avoid an extra boolean function.
The coords argument will allow to use only the subset defined there to do the plots, but not to compare between chains (i.e. creating a plot pair comparing the first and second chain of the same dimension of the same variable). This is going one step further. Moreover, plot_pair does not allow that, therefore, the skeleton will have to be modified in order to include this chain comparison functionality.
Hi @OriolAbril , the functions as parameters makes sense. I was thinking of creating the the function that subtracts and plots the two posteriors in a way that we still pass it data, var_names, coords, and it figures out what to plot, so that it can be interchangeable with all the other types of plots that might be used throughout the grid.
Following your advice, here is a rough attempt. It's missing a lot of the bells and whistles from pair_plot etc, but I think it gets the point across. In addition to pair_plot_extended and plot_dist_diff, I made a helper function for both gen_var_dims_list, that gathers the valid pairs between var_names and coords that will be plotted:
import matplotlib.pyplot as plt
from ..data import convert_to_dataset
from ..plots.plot_utils import purge_duplicates, get_coords, _scale_fig_size
from .posteriorplot import plot_posterior
from .pairplot import plot_pair
from itertools import product
def gen_var_dims_list(data, var_names, coords):
"""Generate list of valid var_names and coords pairings
"""
# If value for key in coords is string and not list of string(s),
# parameter_list does not build properly
for j in coords:
if isinstance(coords[j], str):
coords[j] = [coords[j]]
posterior_data = convert_to_dataset(data, group="posterior")
posterior_data = get_coords(posterior_data, coords)
skip_dims = set()
skip_dims = skip_dims.union({"chain", "draw"})
var_dims_list = []
for var_name in var_names:
if var_name in posterior_data:
new_dims = [dim for dim in posterior_data[var_name].dims if dim not in skip_dims]
vals = [purge_duplicates(posterior_data[var_name][dim].values) for dim in new_dims]
dims = [{k: v for k, v in zip(new_dims, prod)} for prod in product(*vals)]
var_dims = [[var_name, d] for d in dims]
var_dims_list += var_dims
return var_dims_list
def plot_dist_diff(data, var_names, coords, textsize=None, figsize=None, ax=None, **kwargs):
var_dims_list = gen_var_dims_list(data, var_names, coords)
assert len(var_dims_list) == 2, "Too many parameters provided"
(figsize, ax_labelsize, _, xt_labelsize, _, _) = _scale_fig_size(
figsize, textsize, 1, 1
)
if ax is None:
fig, ax = plt.subplots(figsize=figsize, constrained_layout=True)
var_name_0 = var_dims_list[0][0]
var_name_1 = var_dims_list[1][0]
coord_0 = var_dims_list[0][1]
coord_1 = var_dims_list[1][1]
if len(coord_0) == 1:
coord_0_suff = "_" + list(coord_0.values())[0]
else:
coord_0_suff = ""
if len(coord_1) == 1:
coord_1_suff = "_" + list(coord_1.values())[0]
else:
coord_1_suff = ""
diff_DataArray = data.posterior[var_name_0].sel(coord_0) - data.posterior[
var_name_1
].sel(coord_1)
diff_var_name = f"{var_name_0}{coord_0_suff} - {var_name_1}{coord_1_suff}"
data.posterior[diff_var_name] = diff_DataArray
plot_posterior(data, var_names=[diff_var_name], ax=ax, **kwargs)
def pair_plot_extended(data, var_names, coords=None, lower_fun=plot_pair, upper_fun=None, diag_fun=plot_posterior,
lower_kwargs=None, upper_kwargs=None, diag_kwargs=None):
if coords is None:
coords = {}
if lower_kwargs is None:
lower_kwargs = {}
if upper_kwargs is None:
upper_kwargs = {}
if diag_kwargs is None:
diag_kwargs = {}
var_dims_list = gen_var_dims_list(data, var_names, coords)
fig, axes = plt.subplots(len(var_dims_list), len(var_dims_list), figsize=(10, 10))
for i in range(len(var_dims_list)):
for j in range(len(var_dims_list)):
sub_var_name = purge_duplicates([var_dims_list[i][0], var_dims_list[j][0]])
if len(var_dims_list[i][1]) > 0 and len(var_dims_list[j][1]) > 0:
if list(var_dims_list[i][1].keys())[0] in var_dims_list[j][1]:
dict1 = var_dims_list[i][1]
dict2 = var_dims_list[j][1]
sub_coords = {
i: purge_duplicates(list(j))
for i in dict1.keys()
for j in zip(dict1.values(), dict2.values())
}
else:
sub_coords = {**var_dims_list[i][1], **var_dims_list[j][1]}
else:
sub_coords = {**var_dims_list[i][1], **var_dims_list[j][1]}
ax = axes[j, i]
if i < j:
if lower_fun is not None:
lower_fun(data, var_names=sub_var_name, coords=sub_coords, ax=ax, **lower_kwargs)
else:
ax.axis("off")
elif i > j:
if upper_fun is not None:
upper_fun(data, var_names=sub_var_name, coords=sub_coords, ax=ax, **upper_kwargs)
else:
ax.axis("off")
elif i == j:
if diag_fun is not None:
diag_fun(data, var_names=sub_var_name, coords=sub_coords, ax=ax, **diag_kwargs)
else:
ax.axis("off")
plt.tight_layout()In plot_dist_diff, according to the combination of var_names/coords provided, the posteriors of the two parameters are subtracted, and added as a new var_name in data, making the call to plot_posterior simpler (for me)
I maintained the flexibility that the user might provided multiple var_names and want to compare and subtract the posteriors, and that the user knows what they are doing.
So we can have usage like this:
import arviz as az
import matplotlib.pyplot as plt
data = az.load_arviz_data("centered_eight")
az.plot_dist_diff(
data,
var_names=['theta'],
coords={'school': ['Choate', 'Deerfield']},
)
and
az.pair_plot_extended(
data,
var_names=["theta"],
coords={"school": ["Choate", "Deerfield", "Hotchkiss"]},
upper_fun=az.plot_dist_diff,
upper_kwargs={'kind': 'hist'},
lower_kwargs={'plot_kwargs': {'alpha': 0.5}},
)
and
az.pair_plot_extended(
data,
var_names=["theta", "mu"],
coords={"school": ["Choate", "Deerfield", "Hotchkiss"]},
upper_fun=None,
lower_kwargs={'plot_kwargs': {'alpha': 0.5}},
)
Figuring out the lists of all the variables and valid coords is a bit messy here, but it was based off code I found that pair_plot uses.
The labels get a bit messy with so many plots. And (in this example) when a theta is pair_plot-ed with another variable like mu or theta, the coord doesn't get included on the plot.
Is this along the lines of what you were thinking? It's not as quite a simple skeleton as you suggested, but I think it is robust to different types of plots a user might want.
Yes, this definitely follows my line of thought. I will keep playing around with it, to get the most of pairplot and this proposal. It is really good work. I do like the idea of passing xarray datasets, var_names and coords because it will probably allow more options than simply passing the data and the axes. However, it may make the creation of custom plotting functions harder, as it will require understanding xarray and its selection methods.
Do you mind trying this option a little bit and give some feedback? It is extremely similar to the current pair_plot, with some variations:
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.ticker import NullFormatter
from ..data import convert_to_dataset, convert_to_inference_data
from .plot_utils import _scale_fig_size, xarray_to_ndarray, get_coords
from ..utils import _var_names
from .posteriorplot import plot_posterior
from .pairplot import plot_pair
def plot_func_posterior(data, ax, np_fun=np.diff, **kwargs):
data = convert_to_dataset(data)
var_names = [name.replace("\n", "_") for name in data.data_vars]
func_name = np_fun.__name__
xlabel = "{}({},\n{})".format(func_name, *var_names)
data = np_fun(data.to_array(), axis=0).squeeze()
plot_posterior({xlabel: data}, ax=ax, **kwargs)
def plot_pair_extended(
data,
var_names,
coords=None,
combined=True,
lower_fun=plot_pair,
upper_fun=None,
diag_fun=plot_posterior,
lower_kwargs=None,
upper_kwargs=None,
diag_kwargs=None,
figsize=None,
labels='edges',
ax=None,
):
if coords is None:
coords = {}
if lower_kwargs is None:
lower_kwargs = {}
if upper_kwargs is None:
upper_kwargs = {}
if diag_kwargs is None:
diag_kwargs = {}
if labels not in ("edges", "all", "none"):
raise ValueError("labels must be one of (edges, all, none)")
# Get posterior draws and combine chains
data = convert_to_inference_data(data)
posterior_data = convert_to_dataset(data, group="posterior")
var_names = _var_names(var_names, posterior_data)
flat_var_names, _posterior = xarray_to_ndarray(
get_coords(posterior_data, coords), var_names=var_names, combined=combined
)
flat_var_names = np.array(flat_var_names)
numvars = len(flat_var_names)
(figsize, _, _, _, _, _) = _scale_fig_size(figsize, None, numvars, numvars)
if ax is None:
_, ax = plt.subplots(numvars, numvars, figsize=figsize, constrained_layout=True)
for i in range(numvars):
for j in range(numvars):
index = np.array([i, j], dtype=int)
if i > j:
if lower_fun is not None:
lower_fun(
{flat_var_names[j]: _posterior[j], flat_var_names[i]: _posterior[i]},
ax=ax[i, j],
**lower_kwargs
)
else:
ax[i, j].axis("off")
elif i < j:
if upper_fun is not None:
upper_fun(
{flat_var_names[j]: _posterior[j], flat_var_names[i]: _posterior[i]},
ax=ax[i, j],
**upper_kwargs
)
else:
ax[i, j].axis("off")
elif i == j:
if diag_fun is not None:
diag_fun({flat_var_names[i]: _posterior[i]}, ax=ax[i, j], **diag_kwargs)
else:
ax[i, j].axis("off")
if (i + 1 != numvars and labels=="edges") or labels=="none":
ax[i, j].axes.get_xaxis().set_major_formatter(NullFormatter())
ax[i, j].set_xlabel("")
if (j != 0 and labels=="edges") or labels=="none":
ax[i, j].axes.get_yaxis().set_major_formatter(NullFormatter())
ax[i, j].set_ylabel("")There also is a kind of wrapper to plot the 1D posterior of a 2 argument function. Its default is np.diff, which gives the same result as plot_dist_diff, but it can be changed to another numpy function or even some custom function.
In plot_pair_extended, I have added some arguments. The combined argument is used to compare different chains when false, and the labels gives some very rough control over the axis labels and ticklabels.
Some examples:
Default behaviour of plot_pair_extended:
az.plot_pair_extended(
data,
var_names=["theta"],
coords={"school": ["Choate", "Deerfield", "Hotchkiss"]},
figsize=(7,7),
)
Using the wrapper on np.diff as upper_fun:
az.plot_pair_extended(
data,
var_names=["theta"],
coords={"school": ["Choate", "Deerfield", "Hotchkiss"]},
figsize=(7,7),
upper_fun=az.plot_func_posterior,
upper_kwargs={"textsize": 12, "kind":"hist"},
)
Defining a custom plotting function to plot the name of the variables on the diagonal:
def plot_name(data, ax, **kwargs):
name = list(data.keys())[0]
ax.text(.5,.5,name,verticalalignment="center",horizontalalignment="center",**kwargs)
ax.axis('off')
az.plot_pair_extended(
data,
var_names=["theta"],
coords={"school": ["Choate", "Deerfield", "Hotchkiss"]},
diag_fun=plot_name,
diag_kwargs={"fontsize":17},
upper_fun=az.plot_pair,
labels="all",
figsize=(7,7),
)
This is quite far form what you are interested in, but it is basically to try many different arguments. Comparison between different chains:
az.plot_pair_extended(
data,
var_names=["tau"],
combined=False,
diag_fun=plot_name,
diag_kwargs={"fontsize":17},
upper_fun=az.plot_pair,
upper_kwargs={"kind":"kde", "fill_last":False},
lower_kwargs={"plot_kwargs": {"marker":"+","color":"darkblue"}},
labels="none",
figsize=(7,7),
)
This is very nice @OriolAbril ! 👌 So you would be proposing adding both plot_func_posterior and plot_pair_extended to arviZ? I see how plot_func_posterior is being sent a dictionary of the two schools in this case. Could you also show if plot_func_posterior was used alone to create a single difference of posteriors (in this case)?
The customization of labels is very nice. Is there any value of aligning the xlim/ylim for corresponding plots along directions of same variables, or the user would understand that they're not being shown on the same scale (plot_pair and normal plot_posterior in this case)?
It would be nice to reduce the axes grid if only one of the triangles is being shown (and not the diagonal). That can be handled easily while looping through.
Yes, I think that these two could be useful. The plot_name was only for the example. I ended up deciding to pass a dictionary because it can be converted to inference data, which allows to use ArviZ plots, and it is easy to work with, there is only the key which is the label and the item which is the array.
I gave no thought at all to use plot_func_posterior outside pair plot extended, it is quite weird the result as it is now:
data = az.load_arviz_data("centered_eight")
fig, ax = plt.subplots(1,1)
d1 = data.posterior["theta"].sel(school=["Choate"]).to_dict()["data"]
d2 = data.posterior["theta"].sel(school=["Deerfield"]).to_dict()["data"]
az.plot_func_posterior({"Choate":d1, "Deerfield":d2},ax=ax)I am still thinking about the axis. I have some options in mind, but none of them convince me completely. I think it may be useful to have some argument similar to the labels one, so that when plotting a 2d kde and 1d kde, the axis is shared.
The dictionary is definitely a clean usage from within pair_plot_extended. Maybe plot_func_posterior could be updated to also receive data like:
data.posterior['theta'].sel({"school":['Choate', 'Deerfield']})
This could be done by modifying plot_func_posterior a little bit:
def plot_func_posterior(data, ax, np_fun=np.diff, **kwargs):
if isinstance(data, xr.core.dataarray.DataArray):
var_names, data = xarray_to_ndarray(data)
data = np_fun(data, axis=0).squeeze()
else:
data = convert_to_dataset(data)
var_names = data.data_vars
data = np_fun(data.to_array(), axis=0).squeeze()
var_names = [name.replace("\n", "_") for name in var_names]
func_name = np_fun.__name__
xlabel = "{}({},\n{})".format(func_name, *var_names)
plot_posterior({xlabel: data}, ax=ax, **kwargs)So:
data = az.load_arviz_data("centered_eight")
fig, ax = plt.subplots(1,1)
az.plot_func_posterior(data.posterior['theta'].sel({"school":['Choate', 'Deerfield']}), ax=ax)Do you have an idea what other types of functions might be useful to pass to plot_func_posterior?
Sharing the axes is useful, otherwise, hopefully it's obvious to the user when they're not shared (and not labeled). The posterior plots can manage with this if the HPD is displayed.
Do you have an idea what other types of functions might be useful to pass to plot_func_posterior?
I have used the ratio of the inferred variables in some cases.
This is starting to look quite defined! :) Thanks for all the work. I think that now we can start paying attention to presentation and performance. So many conversion from dict to dataset and so on worry me a little.
Okay great.
Further regarding the axes, there are different ways to share/unshare the axes. The problem is quickly/cleanly knowing which ones to share. plot_pair and plot_posterior will be very standard usage, so we can check for their usage and share appropriate plots using those, but not assume anything else about other passed functions, or try to share any other axes.
Maybe it is best to forget about sharing axis. It should not be too difficult to set the limits manually to the same values afterwards if it is really important to maintain this.
@HectorM14 what about using the .sel() (see #669 ) method with inplace=False to pass to the plotting functions (lower, diag...) a whole inference data that contains only 2 variables and the desired coords? Do you have time to work on a PR? Otherwise I will try to do it and I'll let you know once it is good for review, it would be great if you could test it.
Hi @OriolAbril I can hopefully work on the PR this weekend if that works for you. I can try using .sel() as you suggest. Thanks!
Hi @OriolAbril I can hopefully work on the PR this weekend if that works for you. I can try using
.sel()as you suggest. Thanks!
I don't know if the PR adding the .sel() method will be merged though. Check it beforehand.
Got it, when do you think it might be merged? I'll do everything else and wait for that change and try it out.
Probably less than a week
Hi, when we have some time, I think this is something we probably need.
Some functional form to use (at least predefined settings)
lower | diag | upperI think this fell off my radar after waiting for .sel() mentioned above. I can revisit where I left it, or @OriolAbril can finish it too if interested
yeah, no hurry, but I think this is one plot type we are missing.
Basically I would be happy with
lower = scatter diag = posterior upper = stats (cross correlation, mean (vector), sd (vector))
I did some experiments at some point that may help.
I was finally returning to this issue this week, and saw that #1079 is similar. Is this issue still alive or are there still outstanding items to address?
@OriolAbril @aloctavodia @ahartikainen @agustinaarroyuelo
I think the issue is still live, the usecase of adding the marginals on the diagonal, but the idea discussed here covers an even wider range of options. I think it would be a great addition to ArviZ. If plot hierarchy was already in place this would be much easier to handle i think, I am not sure if the rewrite will happen in the near future though.
Okay great, I'll come back with an update this weekend!
I've been working on this, and either some things have changed in the ten months since I raised this issue, or I'm thinking about it differently. As part of this issue/PR, I think that pair_plot_extended (or whatever this function ends up being named) will be a really flexible plot with any combination of upper/lower triangles with or without diagonal, with standard or custom plot types. When @OriolAbril and I left off, pair_plot_extended was flexible, but at its base, used pair_plot as default.
I've been thinking that it would make a lot of sense to build pair_plot_extended as described above, and when pair_plot is called to make a grid of axes (numvars > 2), it just calls pair_plot_extended (to get a lower triangle, or lower triangle + diagonal). I think that will limit code reuse, and simpler plots like pair_plot can just request a simpler plot from pair_plot_extended. From a user's perspective, the API to pair_plot would be the same.
I'd be curious to hear what you think, and may investigate this direction unless you have some objections.
@OriolAbril @aloctavodia @ahartikainen
At least for now I'd keep plot pair and plot pair extended as completely independent. I would be open to the merge eventually once plot pair extended is up and running, we can be sure it will handle all plot_pair options and we have run some benchmarks. plot pair is an extremely common plot, with not much but some level of optimization to its specific case, and plot pair extended may have to renounce to some of it in order to be flexible enough. 🤔
If you feel you are using too much the same code we can think about modularizing the code and creating some functions in plot utils with the parts common to plot pair and plot pair extended.
Does this sound sensible?
Sure that sounds good @OriolAbril . We'll see what it looks like in the end. Just wanted to pass along the thought before I got too deep
any update on this?
@aloctavodia I'm back on it! Just catching up with all that has changed in the meantime.
Hi @aloctavodia and @OriolAbril I'm back! I'm dedicating time this week (or longer) to finally settle this issue.
I have an updated branch of our progress here: https://github.com/hectormz/arviz/blob/plot-pair-extended/arviz/plots/pairextendedplot.py
I'm in the process of updating the branch to match arviz changes that have happened since I worked on it last.
Most recently, we had planned to implement more use of .sel() when passing prior/posterior data to the upper/lower/diagonal plotting functions. There are two issues that seem to come up:
We are currently using flat_var_names like plot_pair to combine the variable name and coord/dimensions. This is in part why we are passing the data to the lower/upper functions as a dictionary, because the flat variable names don't exist in the original posterior. Even if we use the pattern of creating a list of plotters like plot_pair does, we have to update the names and it is still a dictionary of data.
x and y, and pass var_name & coord arguments (and let the second plot functions take care of making flat_var_names), we come across the issue presented below.The other issue I've come across is that the ordering of x and y axis data seems to be implicit in the arviz plot functions that I've looked at. plot_pair_extended makes the most sense with specific variable/coords on x or y axis for each plot.
This is currently achieved just by the order that the keys appear in the dictionary we send (which would not have been maintained in previous python versions). So even if we were to send a filtered posterior, with a list of appropriate var_names and coords, the second plot functions may not plot in the right order. This can be taken to an extreme if we consider one of the plots above might be comparing chain 0 from tau with Choate chain 1 from theta (in other words, multiple variables and multiple coords levels).
What are your thoughts?
Am I wrong about how the arviz plotting functions choose what goes on x vs y axes? Or is there a way to force it?
Not sure I follow the issue. It might not be very intuitive but the order in which variables are plotted is deterministic and can be modified. There are some examples in https://python.arviz.org/en/latest/user_guide/label_guide.html. Also ArviZ supports only 3.7+ python versions, so we can assume dicts are ordered (and IIRC we should drop 3.7 in next version to follow NEP advise).
If that does not address the concerns, could you maybe share an example to see if I get an idea?
Also, super happy to hear back from you :)
Thanks @OriolAbril ! This long unresolved Issue/PR has brought me shame, which I hope to erase!
Thanks for the pointer on the labels! I think that clears some things up for me. My concern was that we wanted to use fewer dictionaries:
I think that now we can start paying attention to presentation and performance. So many conversion from dict to dataset and so on worry me a little.
what about using the .sel() (see https://github.com/arviz-devs/arviz/pull/669 ) method with inplace=False to pass to the plotting functions (lower, diag...) a whole inference data that contains only 2 variables and the desired coords?
But I think I have the info I need about labels to try to clean up passing the filtered and sorted InferenceData to the plotting functions. I'll see if there are any edge cases that don't work.
Thanks for the guidance @OriolAbril !
@OriolAbril one more question. When we first started this, plot_pair() had an argument combined, which when set to False, prevented the chain coords from being combined and you could generate a pair_plot that looked at the different chains of variables. combined is now gone and with the use of combine_dims, draw and chain are always combined (and others may be added).
Is that right, or is there another way to look at chains? I wanted to make sure this function has similar features as plot_pair, and prevent plotting of individual chains if that is the right behavior. I'm indifferent to looking at individual chains, but you demonstrated it in one of your demos: https://github.com/arviz-devs/arviz/issues/662#issuecomment-492042419
I might have added combined to the draft of plot_pair_extended :sweat_smile:, I am not sure it was ever available in plot_pair. We can leave it out as it will most probably make things easier, no need to keep that example as something we'd want to be possible
Aha! Sounds good
Still more to do, but this https://github.com/hectormz/arviz/blob/419f13dac79cbf1b9238abf8d3668322ad7f7eb6/arviz/plots/pairextendedplot.py#L279 passes a filtered xarray dataset that keeps variable and dim order.
This commit used the plotter pattern used in the normal plot_pair:
https://github.com/hectormz/arviz/blob/968f5dc496592dd3d6ab1c30b3ac88d4fc38bad6/arviz/plots/pairextendedplot.py#L222
This looks great! I love it because it only takes care of data and subplot organization and delegates all data processing and actual plotting to the other functions, therefore it will be compatible with any existing arviz function (as long as it plots the two variables in a single subplot) or any new function that takes inferencedata input. :smile:
Exactly! I need to try this out with some different plot types and see if there are any that misbehave or break in a way that isn't obvious to the user.
Hi @OriolAbril another update: I actually updated how data is sent to the plotting functions. I'm now sending a partially filtered InferenceData (with var_names and coords etc.) for the sake of the arbitrary plotting functions, since that is what they expect if you want to be able to plot divergences etc.
Ideally, it would be nice to have the same infrastructure for matplotlib and bokeh, where a numpy grid of axes is generated for each backend, and ax[j,i] is passed to each plot function. I tried this, but had an issue with bokeh 🤷♂️
I think there's still more work to be done, but might open a WIP PR soon to make it easier for you to follow along.
I wanted a way to efficiently compare multiple marginal posteriors in PyMC3/ArviZ like in Figure 9.10 from Kruschke's book: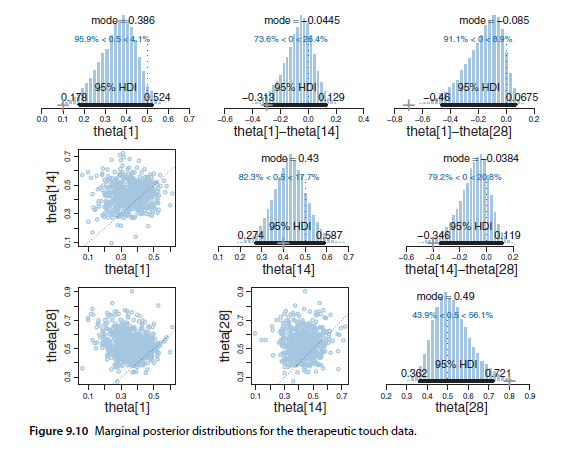
This is especially the case when using vectorized parameters in a model, and I'd like to compare many/all of them. If I have two, creating a
pm.Deterministicdifference isn't bad.I searched PyMC3/ArviZ documentation and examples, and didn't seem to find anything that fit this need. Forest plots give similar answers, but comparing HPDs of two parameters is not the same as looking at the HPD of their difference.
I created a function to plot the difference in marginal posteriors.
Here's the combined forest plot for the same trace: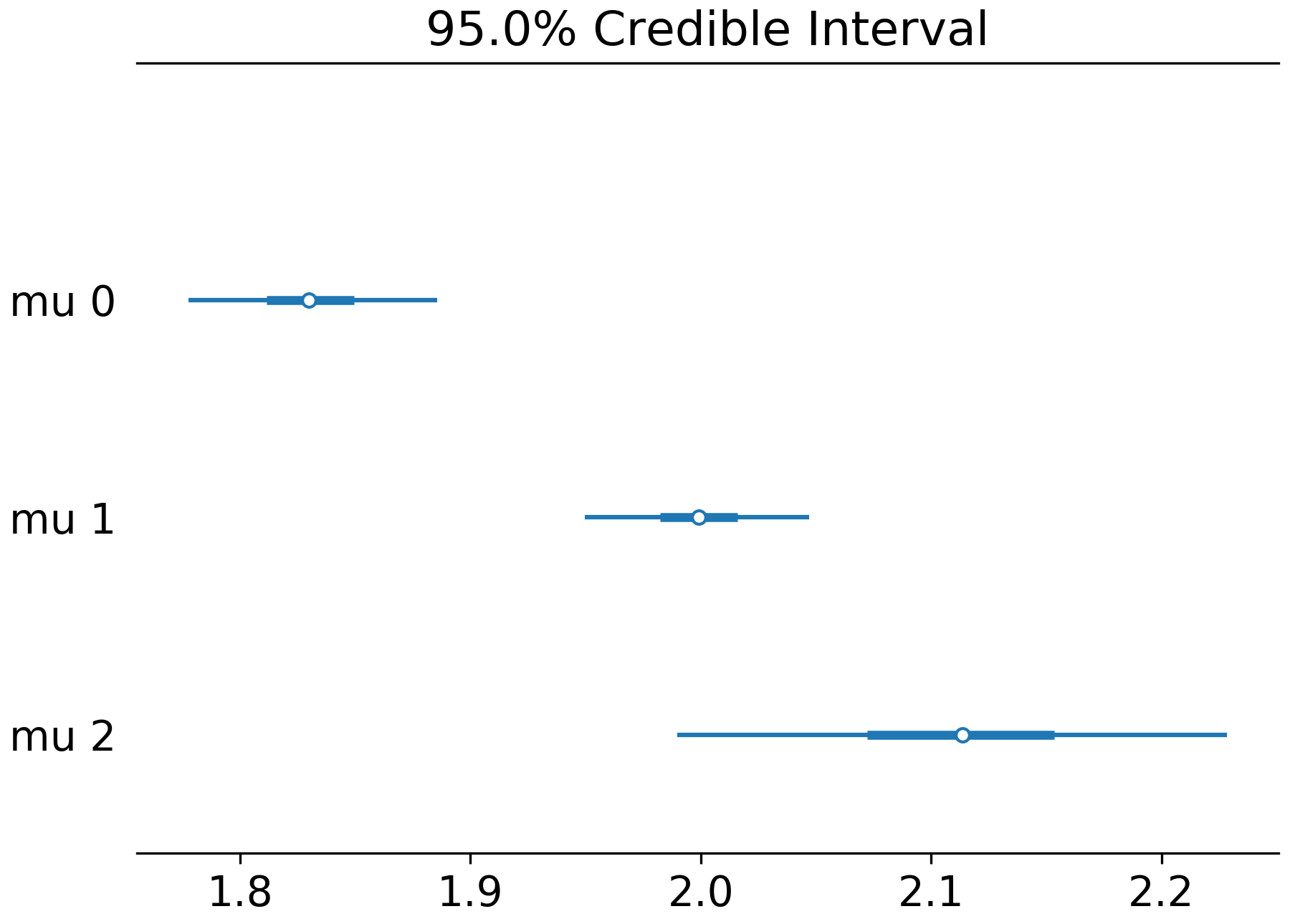
I didn't care about recreating the scatter plots, but the function could be modified to faithfully recreate the original figure: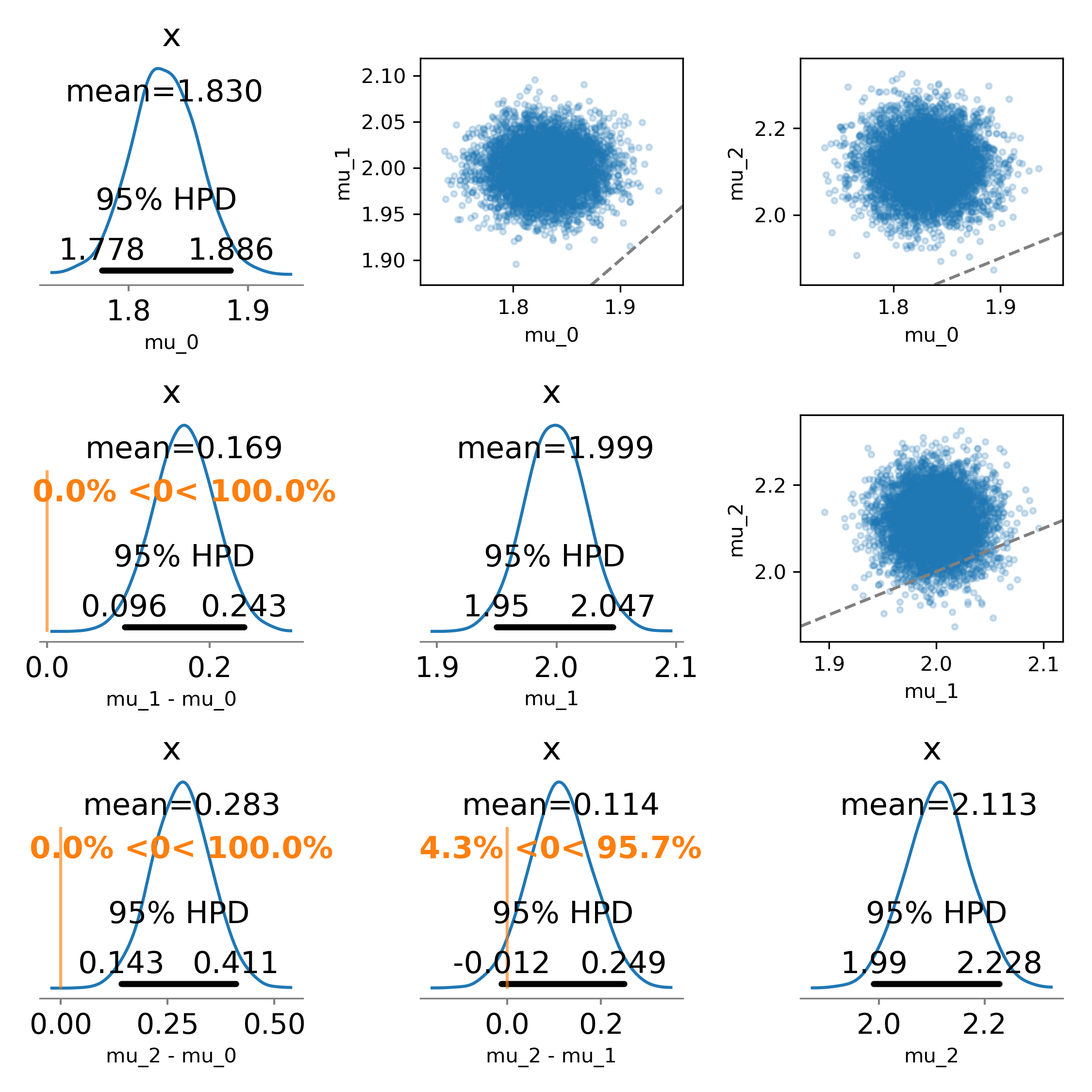
The results (and interpretations) may be different from what you'd get from a forest plot, depending on the data and parameters.
My function assumes that only one parameter would be compared at a time, and assumes that the parameter vector is a reasonable length. It's a little hackish, and assumes a PyMC3 trace for data.
Is this something worth adding to arviZ? Is there any reason that these types of plots are invalid or shouldn't be encouraged? If there's interest, I'd be willing to build this into a PR to add to arviZ (and PyMC3 plotting).