Hi there Chris!
Thank you for spotting the bug.
- I made the fix to the feature hashing.
- The erasing of the base feature is an optional parameter. As I ran some benchmarks it turned out that erasing the base feature reduced the predictive power of the features.
- This change now effects the tree features file and every model class which was effected downstream. (Role2vec, Graph2vec, GL2Vec).
If you upgrade Karate Club to 1.0.11 and modify your testing script and set the "erase_base_features" parameter to false our results should match.
I am happy that you like the package. If you end up using it in a paper please cite the relevant paper (it was accepted in the KDD MLG Workshop and it is also under review for CIKM 2020).
If you are satisfied with this please feel free to close this issue.
Hello,
I noticed an error in the WeisfeilerLehmanHashing class from utils.treefeatures.py which is used by Graph2Vec and Gl2vec for building the list of nodes rooted subtrees of the each graph up to the number of WL iterations (argument: wl_iterations).
I implemented a correction of WeisfeilerLehmanHashing and show you hereafter a comparison to demonstrate the problem.
reproducible code
It (i) generates a simple chain graph (ii) Run the WL algorithm implemented in your original code and print the WL features hash codes outputs. (iii) Run the corrected algorithm and print the WL features hash codes outputs. (iv) Print the WL nodes rooted trees associated with the WL features hash codes (they are in the same order).
Here is the output of this script: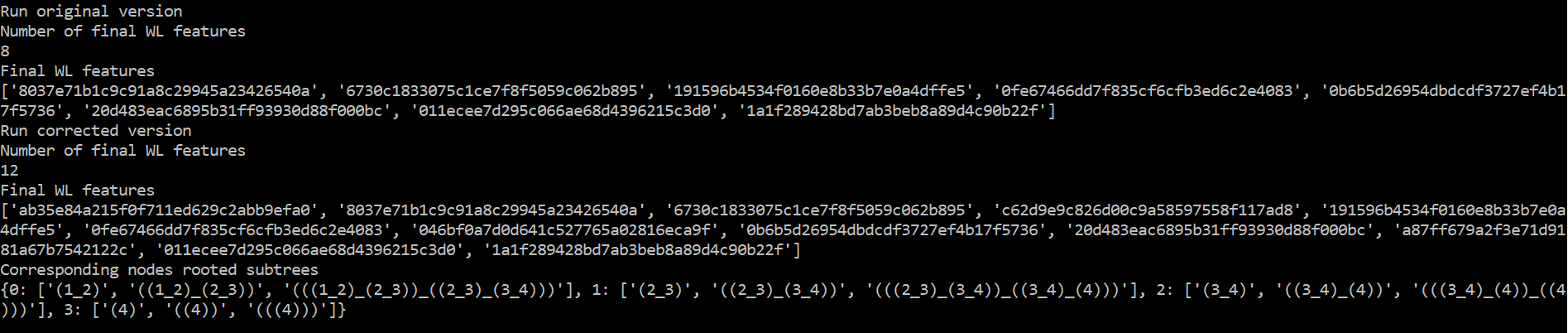
Explanation
Indeed, you can see first that the original code only produces 8 (4x2) WL features hash codes instead of 12 (4x3) in the corrected version. The algorithm must produce one WL feature per node and iteration.
You can also see that 8 codes among the corrected output match the non-corrected output. Now looking at the output of WL.all_subtrees, whose order match the features in WL.get_graph_features(), you can see that the codes that were lacking in the non-corrected version correspond to the first WL iteration.
Summary
The problem of the original code is that it only keep the WL features of the 2 last iterations. I hope that this is clear enough. Of course you can use directly the corrected version. I kept track of the changes and highlighted those that I only use for the trees strings construction.
Could you please apply the changes to the package? I'm using it for research and I think it would be much easier for everyone that I directly refer to your package rather than adding a corrected code in a personal repo.
Anyway, thank you for this useful package =)