Hi @whitleyo , it is an interesting problem. Based on the PCA results, the separation is more clear in some data types than in others, so it is not be surprising if MOFA does not capture a factor that is common across all data types. But it should definitely capture a factor that separates the groups that you are considering. Can you check if this is the case?
P.S. Please upgrade to MOFA2 (https://github.com/bioFAM/MOFA2), it is a better software and we are no longer maintaining this repository. Also, feel free to join the Slack group where we can have a more interactive discussion.
Best, Ricard.
Hi,
I've tried MOFA with several datatypes, some having ~50 samples while others have 15 or 22 samples.
Here's an overview of the data:
RNA.vst = vst transformed RNA-seq data DNAm = DNAm m-values metab_annot_extract = metabolites, cell extract, annotated metab_annot_secreted = metabolites, cell secretion, annotated metab_unannot_extract = metabolites, cell extract, unannotated crispr_qBF = quantile normalized Bayes Factors from CRISPR screens similar to that in Hart et al. 2015, but using a smaller library.
I ran MOFA with the following training options on this data (20 other models were run, most producing similar results, none having a common axis of variation shared between all datatypes)
The resulting model has the following explained variance: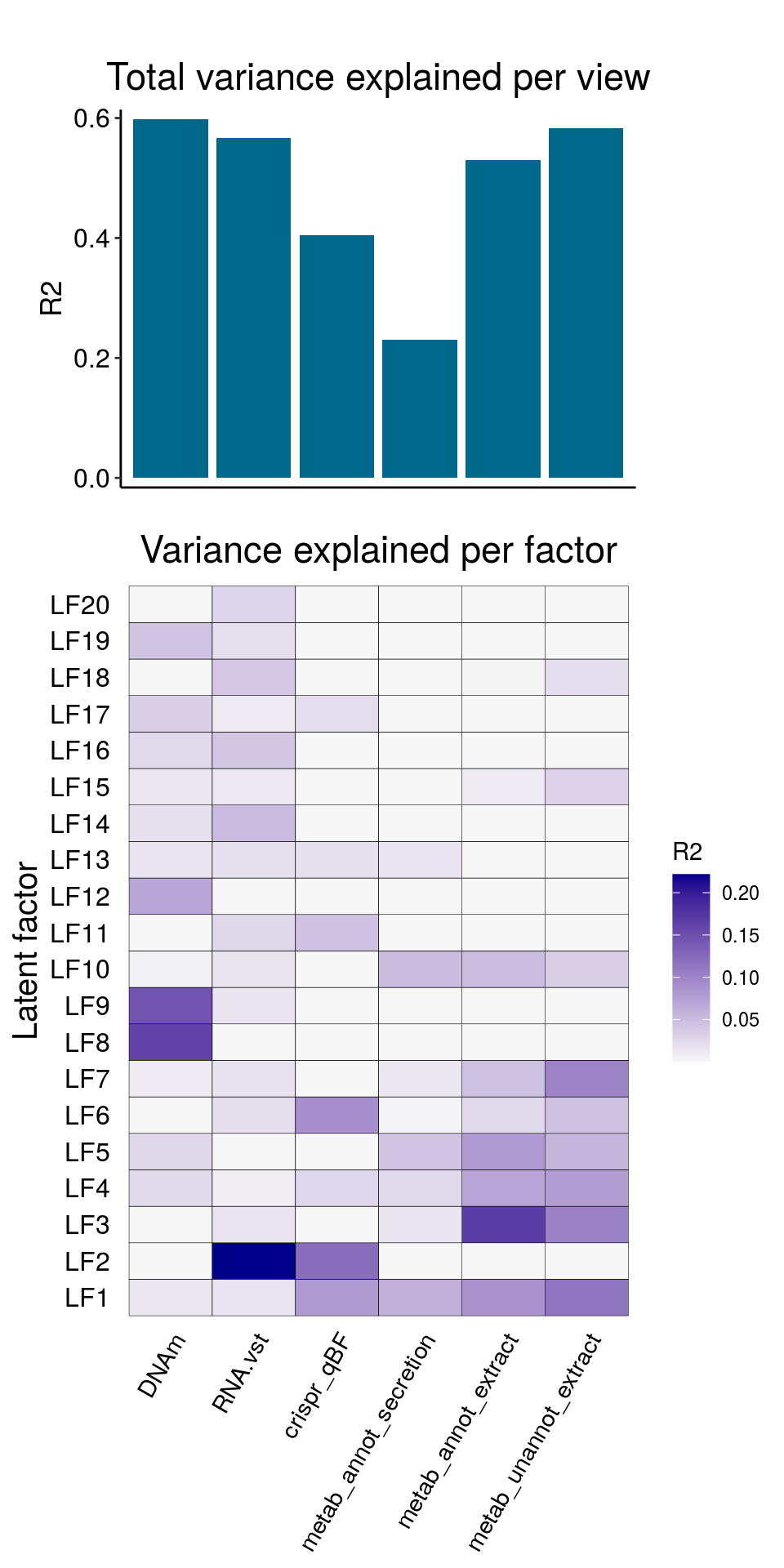
and correlattion between factors: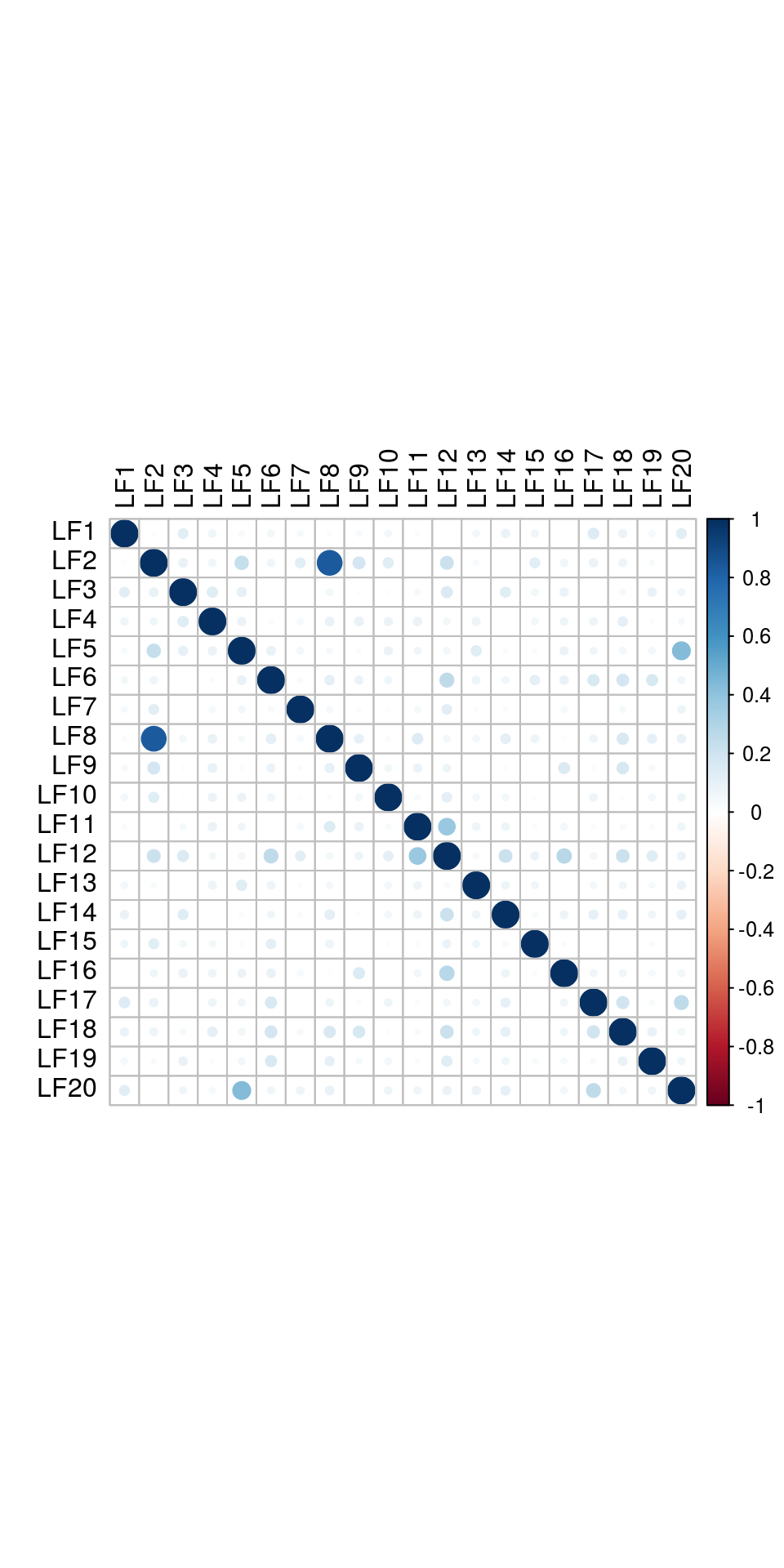
The results would seem to imply that metabolomics data do not share a common axis with the RNA-seq and DNA methylation data. When I run PCA on eahc of the data matrices as input to MOFA individually however, I get clean or relatively clean separation of clusters identified in RNA-seq data in each datatype:
RNA-seq: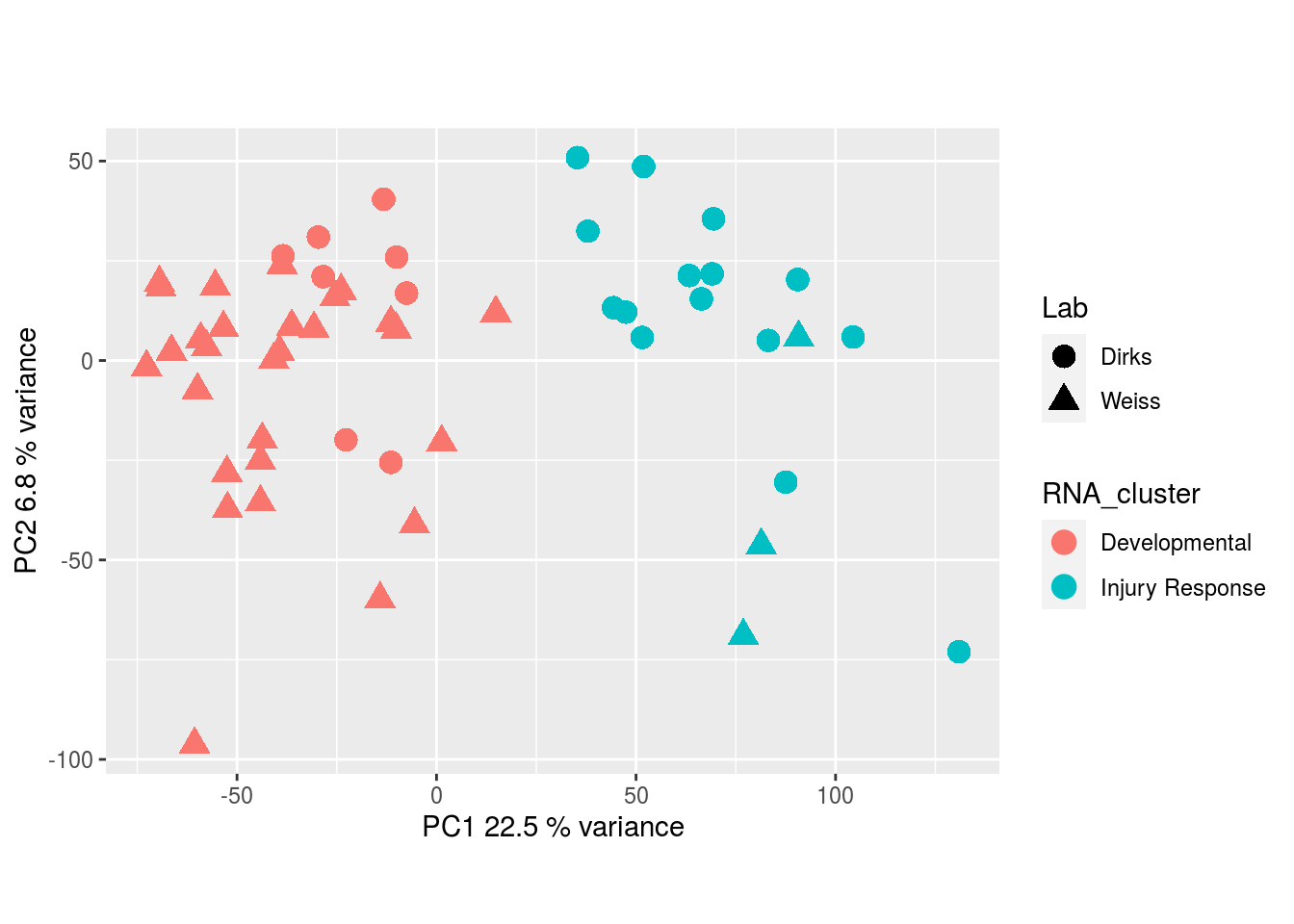
DNA methylation: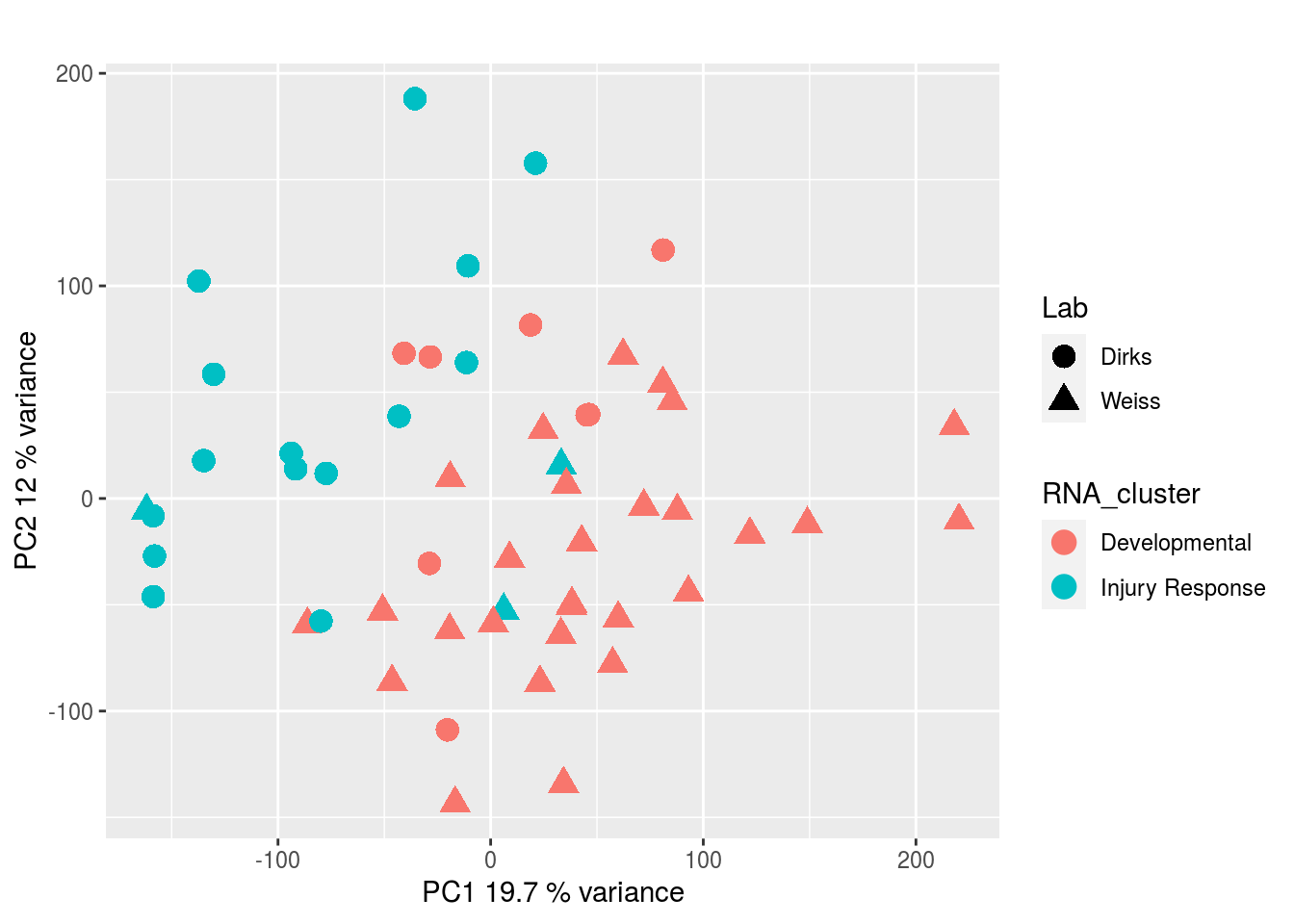
CRISPR Screen: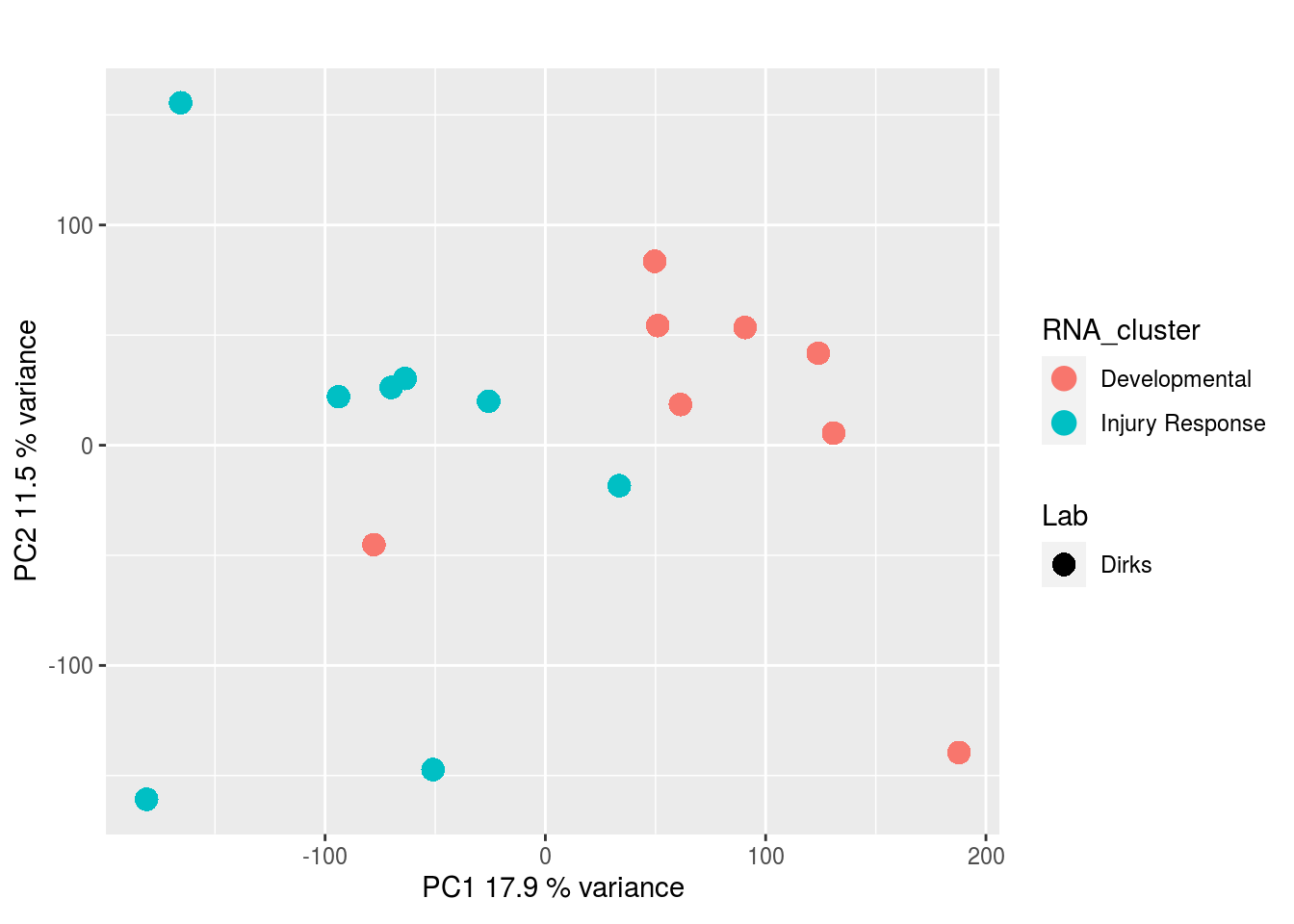
Metabolites, Annotated Cell Extract: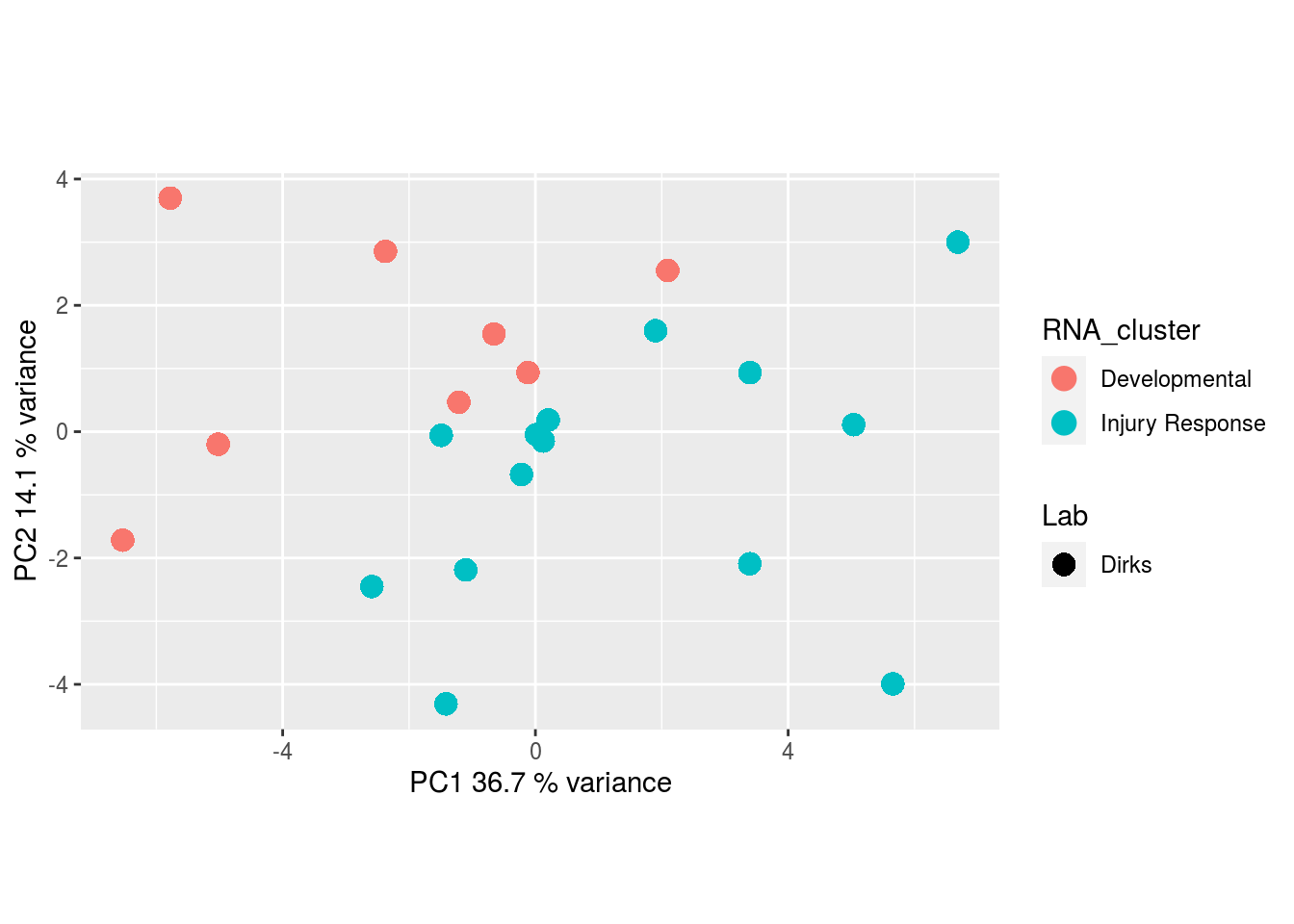
Metabolites, Unannotated Cell Extract: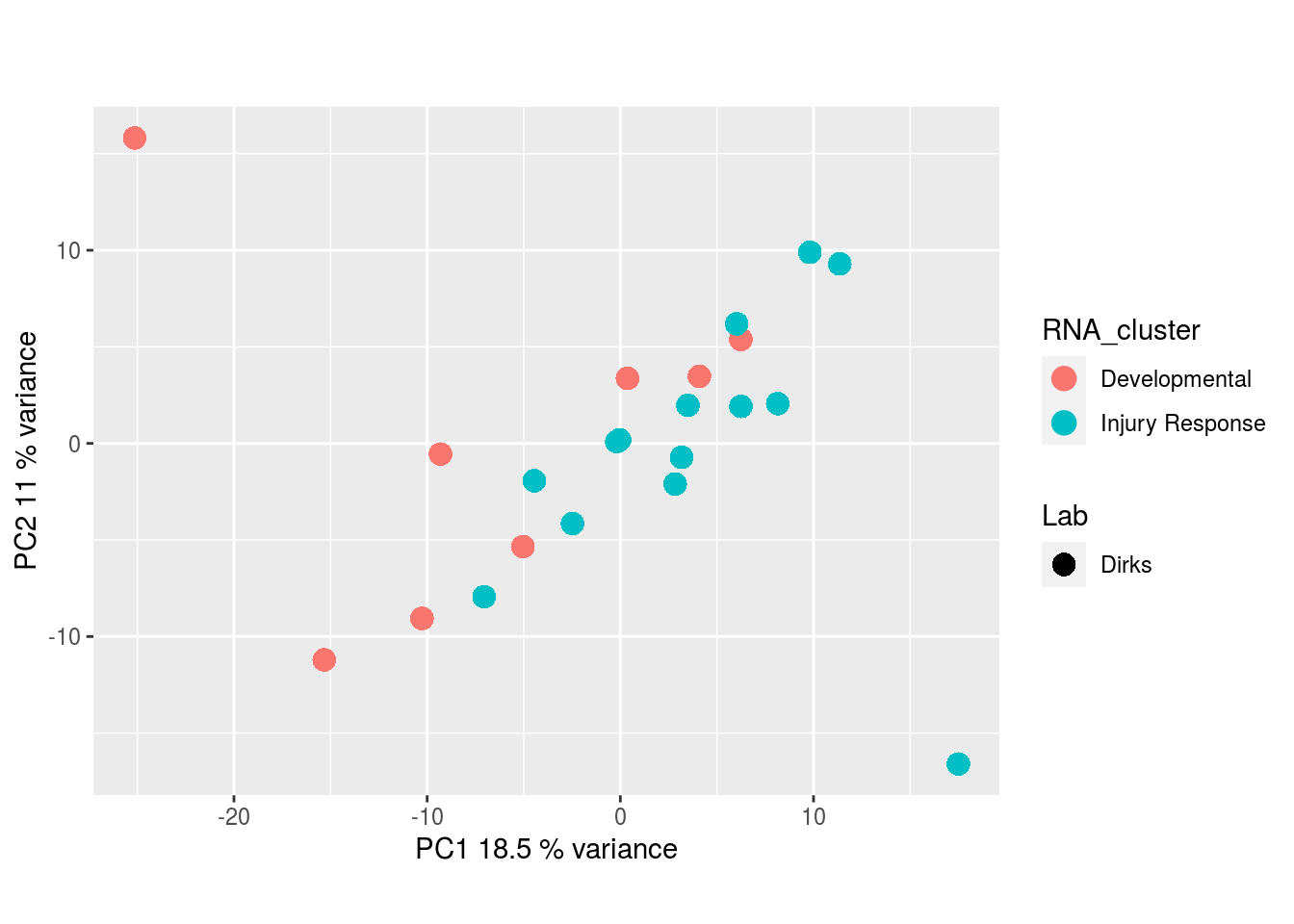
Metabolites, Annotated Secretion: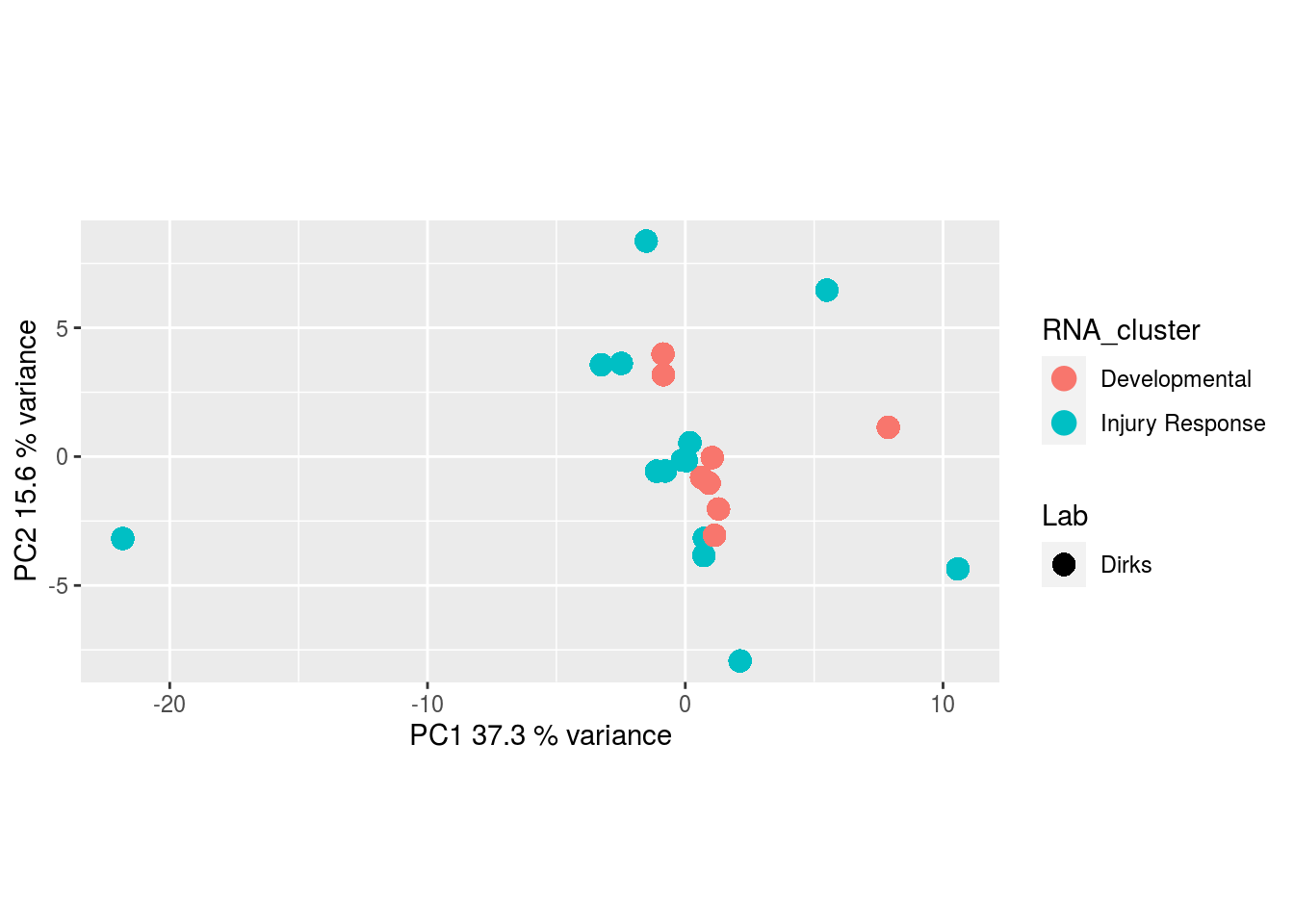
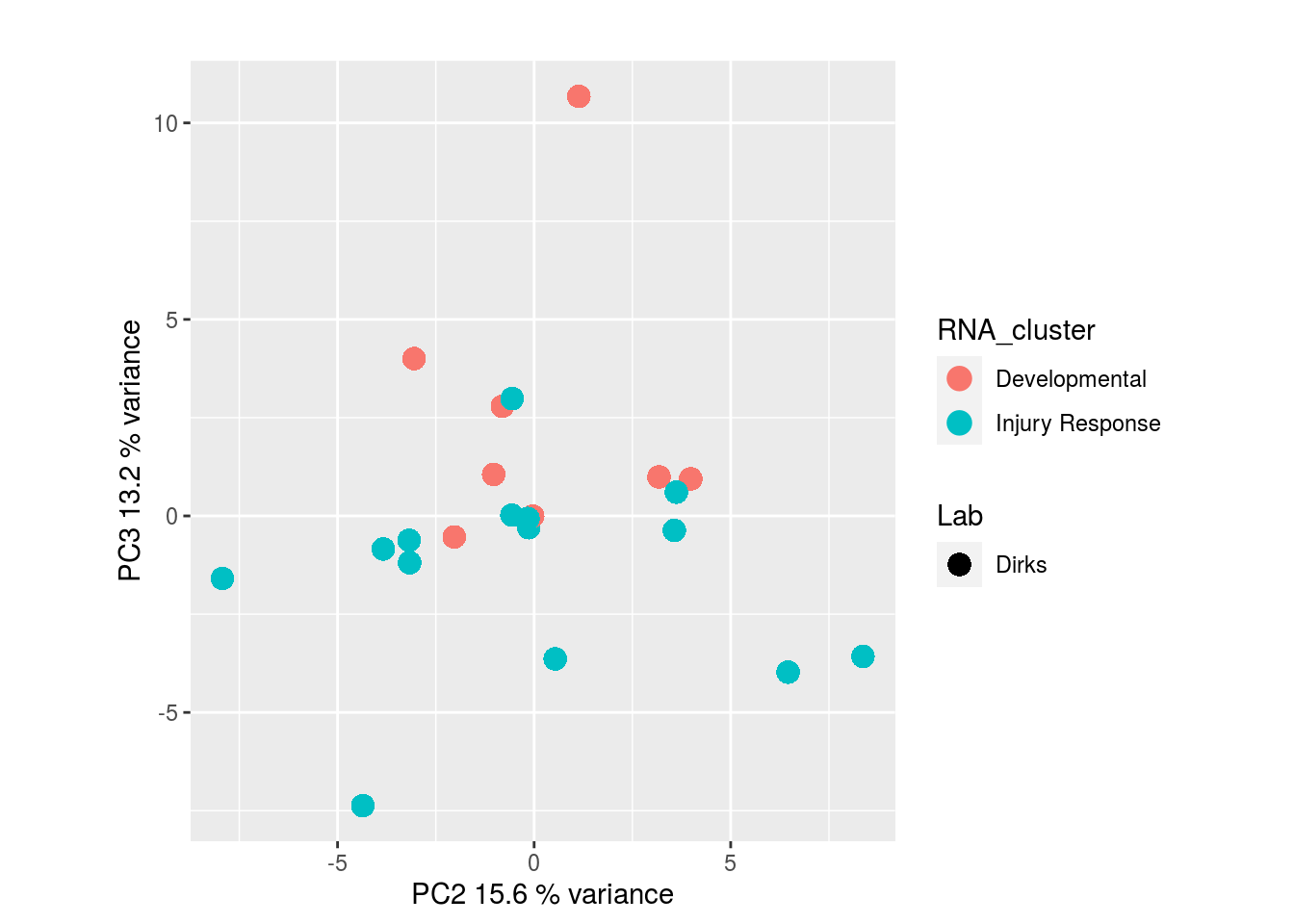
I would have expected a common factor to be found for all datatypes, but I'm wondering if there's too much missing data here.
Session Info: