@gradyrw , as it happens we were looking at marginalization today, but it's been a while since I did a deep-dive - and the students that helped create this are all gone. I am prepared to take a deeper look if you could create a some unit tests that fail. I just pushed a branch fix/iSAM2 which you could create a PR against from a fork, if you're willing.
Description
I have been trying to use ISAM2's marginalizeLeaves function and encountered a few issues, particularly on larger problems. I've tracked the issues down as best I can and I believe there are 3 different bugs in the marginalizeLeaves function which are causing problems:
There is a missing
nullptrcheck which is required in cases where a leaf is also a root (e.g. marginalizing the only node in a graph).The vertical block matrix of the clique's conditional is not getting updated properly in cases where the entire clique is not marginalized
The logic for splitting a clique assumes that keys are ordered in a way such that all the marginalized keys appear before any keys that are being kept. This is not always true.
I think there are simple solutions for the first two issues, but I'm not sure about the final one.
Steps to reproduce
I'm using this code to force the keys that I want to marginalize to be leaves and to then update ISAM2 and marginalize:
I've adapated this code from IncrementalFixedLagSmoother.cpp and it appears to work fine (I haven't encountered any errors from trying to marginalize non-leaf nodes). Next I've got the following helpers for creating an ISAM2 object and defining noise models for factors:
Issue 1: Missing nullptr check
To reproduce this error, just create a factor graph with a single variable and then try to marginalize that variable. Doing so results in a segfault:
I've tracked this down to line 556 in ISAM2.cpp, the parent of the clique is dereferenced without checking if it is null:
In this case the parent of the clique will be null since the node is a root. This is easy to fix, since we are getting rid of the entire tree it's not necessary to keep track of any marginal factors and just adding a null pointer check before pushing the marginal factor works:
Issue 2: Incorrect block matrix update
This issue is harder to trigger and only manifests for larger graphs that have undergone repeated marginalizations. I've tried to make the smallest possible example here. First, create a factor graph with pose variables layed out in a 2D grid. Each variable has a prior factor placed on it and then between factors connect each node to its neighbors:
Then, one by one, we randomly select a variable, marginalize it, and calculate the best estimate:
Make sure that the seed is set to 1234, otherwise results will differ. With that seed this eventually throws an
IndeterminantLinearSystemerror, which it shouldn't be doing. We have a prior on each variable which should constrain the solution. Looking at things a little closer, it throws theIndeterminantLinearSystemerror while working near variable 5, right after it marginalized key 25 from the clique:(25,5,3 : 2,7,11,17,22,26,33,35,43,45,54)If we look at what happens while we are marginalizing key 25, it looks like there are issues with how the conditional's matrix is being updated (
cg->matrixObject()). Lines 634-636 are where this update happens inISAM2.cpp. Immediately before that update, the block matrix for the troublesome clique has the following state:We want to chop off the first 6 rows and first 6 columns of this matrix (since the variable we are marginalizing is a Pose3 type). However, what happens instead is that we chop off the first 6 rows while the columns remain the same. This is the state of the block matrix after line 636:
which is a singular matrix and is the cause of the
IndeterminantLinearSystemerror. The issue is that whilerowStart_gets incremented,firstBlock_only gets set via:on line 635. This isn't correct (although it works if firstBlock() is zero). Instead, firstBlock() should get incremented by
nToRemove. We can do this be changing 635 to:which results in the following block matrix after the update:
which appears to be correct. After this change the
IndeterminantLinearSystemerrors are gone and this part of the test passes.Issue 3: Incorrect ordering assumption
The next issue comes when trying to marginalize the variable with key 3. This variable is in the clique:
(5,3,2,1 : 7,10,17,21,22,26,33,35,43,45,54).After this marginalization call
calculateBestEstimategives the following error:The problem is that the variable with key 3 was not actually removed during the call to marginalizeLeaves. This check on line 631:
sets
nToRemoveas zero sincecg->keys()[0] = 5is not a key that we want to remove. It's simple enough to change this so thatnToRemoveis calculated correctly, but even if we havenToRemove = 1the result will be that variable 5 will be removed instead of variable 3 (or even worse, some parts of variable 5 could be removed or variable 5 and some parts of variable 3).One possible way to fix this is to re-order keys in the clique as we are marginalizing them. I think that replacing line 631 with something like this could work:
I've gotten this work for the case where all variables have the same size, however the general case is a little trickier. The problem is that when you swap matrix blocks you have to shuffle the blocks up or down in order to preserve the upper triangular structure of the matrix. This involves copying data and could wind up being expensive.
I'm hoping that there might also be an easy way to re-order the keys/matrix in the conditional that I'm missing. When I made the modification to swap matrix blocks, the test was able to finish and the results all appeared correct. I'm happy to put up a PR with these changes, but would like some input on the best way to solve this last problem first.
Additional Details
This is the state of the Bayes Tree immediately before the failure described in issue 2:
This is the state of the Bayes Tree immediately before the failure described in issue 3: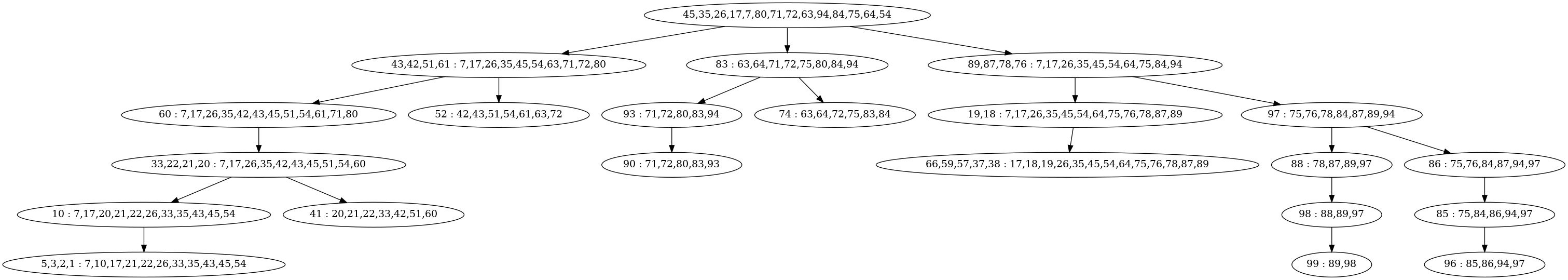
Here is the full code for reproducing these results: