I think the less bins you use the greater txnE you should set. Because larger bins leads to fewer CNA. Greater txnE value gives the model more "stable" copy number change. More detail in the article Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors.
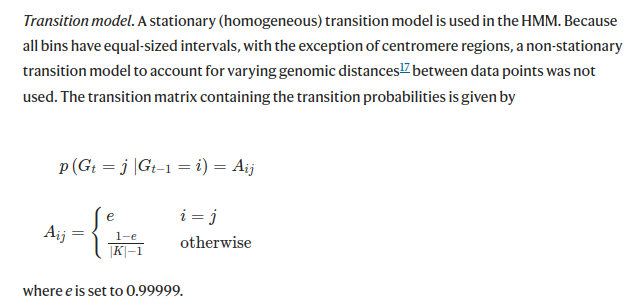
Is there a formula or ratio of how the txnE and txnStrength parameters should relate to the bin width? Should these always kept at default value or is there a rule of thumb for this parameter?
Quote from snakefile: