Hi Liurchen ,
Can you share the config file for test run? This repository does not have the config file for testing.
Closed Liurchen closed 2 years ago
Hi Liurchen ,
Can you share the config file for test run? This repository does not have the config file for testing.
Hi CHENG, thanks for sharing the code. I am running the code on the Combined Healthy Abdominal Organ Segmentation dataset. While after training, the result of the evaluation is quite different from that represented in the paper. I am wondering whether do I have some misoperation. Here my steps are shown below: At first, I downloaded datasets from https://chaos.grand-challenge.org/Download/. And I got datasets like this:
Then as mentioned in the README.md file, I put the MR folder into
Self-supervised-Fewshot-Medical-Image-Segmentation/data/CHAOST2. In the data pre-processing, first I run./data/CHAOST2/dcm_img_to_nii.shand./data/CHAOST2/png_gth_to_nii.ipynpto convert dicom images to nifti files. After that, I got a folder namedniis/T2SPIR, it consists of images in.nii.gzformat.Then I run
./data/CHAOST2/image_normalize.ipynbto do the pre-processing of the download images, and got normalized image folderchaos_MR_T2_normalized. Next I run./data/CHAOST2/class_slice_index_gen.ipynbto build class-slice indexing for setting up experiments. Then to generate pseudo label, run./data/pseudolabel_gen.ipynb, after all this done, the folder I got is shown below:Then I started to train the model by running
./examples/train_ssl_abdominal_mri.shand here my config file is shown below:""" Experiment configuration file Extended from config file from original PANet Repository """ import os import re import glob import itertools import sacred from sacred import Experiment from sacred.observers import FileStorageObserver from sacred.utils import apply_backspaces_and_linefeeds from platform import node from datetime import datetime sacred.SETTINGS['CONFIG']['READ_ONLY_CONFIG'] = False sacred.SETTINGS.CAPTURE_MODE = 'no' ex = Experiment('mySSL') ex.captured_out_filter = apply_backspaces_and_linefeeds source_folders = ['.', './dataloaders', './models', './util'] sources_to_save = list(itertools.chain.from_iterable( [glob.glob(f'{folder}/*.py') for folder in source_folders])) for source_file in sources_to_save: ex.add_source_file(source_file) @ex.config def cfg(): """Default configurations""" seed = 1234 gpu_id = 0 mode = 'train' # for now only allows 'train' num_workers = 4 # 0 for debugging. dataset = 'CHAOST2_Superpix' # i.e. abdominal MRI use_coco_init = True # initialize backbone with MS_COCO initialization. Anyway coco does not contain medical images ### Training n_steps = 100100 batch_size = 1 lr_milestones = [ (ii + 1) * 1000 for ii in range(n_steps // 1000 - 1)] lr_step_gamma = 0.95 ignore_label = 255 print_interval = 100 save_snapshot_every = 25000 max_iters_per_load = 1000 # epoch size, interval for reloading the dataset scan_per_load = -1 # numbers of 3d scans per load for saving memory. If -1, load the entire dataset to the memory which_aug = 'sabs_aug' # standard data augmentation with intensity and geometric transforms input_size = (256, 256) min_fg_data='100' # when training with manual annotations, indicating number of foreground pixels in a single class single slice. This empirically stablizes the training process label_sets = 0 # which group of labels taking as training (the rest are for testing) exclude_cls_list = [2, 3] # testing classes to be excluded in training. Set to [] if testing under setting 1 usealign = True # see vanilla PANet use_wce = True ### Validation z_margin = 0 eval_fold = 0 # which fold for 5 fold cross validation support_idx=[-1] # indicating which scan is used as support in testing. val_wsize=2 # L_H, L_W in testing n_sup_part = 3 # number of chuncks in testing # Network modelname = 'dlfcn_res101' # resnet 101 backbone from torchvision fcn-deeplab clsname = None # reload_model_path = None # path for reloading a trained model (overrides ms-coco initialization) proto_grid_size = 8 # L_H, L_W = (32, 32) / 8 = (4, 4) in training feature_hw = [32, 32] # feature map size, should couple this with backbone in future # SSL superpix_scale = 'MIDDLE' #MIDDLE/ LARGE model = { 'align': usealign, 'use_coco_init': use_coco_init, 'which_model': modelname, 'cls_name': clsname, 'proto_grid_size' : proto_grid_size, 'feature_hw': feature_hw, 'reload_model_path': reload_model_path } task = { 'n_ways': 1, 'n_shots': 1, 'n_queries': 1, 'npart': n_sup_part } optim_type = 'sgd' optim = { 'lr': 1e-3, 'momentum': 0.9, 'weight_decay': 0.0005, } exp_prefix = '' exp_str = '_'.join( [exp_prefix] + [dataset,] + [f'sets_{label_sets}_{task["n_shots"]}shot']) path = { 'log_dir': './runs', 'SABS':{'data_dir': "/mnt/c/ubunturoot/Self-supervised-Fewshot-Medical-Image-Segmentation/data/SABS/sabs_CT_normalized" }, 'C0':{'data_dir': "feed your dataset path here" }, 'CHAOST2':{'data_dir': "/home/rc/Documents/Self-supervised-Fewshot-Medical-Image-Segmentation/data/CHAOST2/chaos_MR_T2_normalized/" }, 'SABS_Superpix':{'data_dir': "/mnt/c/ubunturoot/Self-supervised-Fewshot-Medical-Image-Segmentation/data/SABS/sabs_CT_normalized"}, 'C0_Superpix':{'data_dir': "feed your dataset path here"}, 'CHAOST2_Superpix':{'data_dir': "/home/rc/Documents/Self-supervised-Fewshot-Medical-Image-Segmentation/data/CHAOST2/chaos_MR_T2_normalized/"}, } @ex.config_hook def add_observer(config, command_name, logger): """A hook fucntion to add observer""" exp_name = f'{ex.path}_{config["exp_str"]}' observer = FileStorageObserver.create(os.path.join(config['path']['log_dir'], exp_name)) ex.observers.append(observer) return configAfter training, I have got some snapshot of the model:
Then I run
./examples/test_ssl_abdominal_mri.shto test the model, and below is my result:As is shown in the picture, the dice score is about 0.8, while in the paper, it shoule be around 80, it's quite different. So I wonder if there are some mistakes during my experiment? And how could I fix it and get ideal results? I would deeply appreciate your reply.
Hello, the data in the paper is a percentage, which means that 0.8 is represented by 80%, and the "%" is omitted in the table. Your results are no problem.
Hi, can you run CT test successfully? MRI test have no error, but when I test CT, the following problems arise:

Hello, I also encountered this problem at the time and then solved it; the reason is that there is a problem with the code in the "Synapse_abdominal_classmap.ipynb" file that processes the SABS data set. The original code is:
Please replace this part of the red box code (as shown in the figure below) directly with this part of "class_slice_index_gen.ipynb" in CHAOST2 dir, and then re-run to generate superpixel label for CT images.
The details is: the number of slices read by "nio.read_nii_bysitk" is in the first dimension, that is "n_slice=shape[0]". According to this understanding, the label can be processed smoothly. Good luck!
------------------ 原始邮件 ------------------ 发件人: "cheng-01037/Self-supervised-Fewshot-Medical-Image-Segmentation" @.>; 发送时间: 2021年6月2日(星期三) 晚上6:24 @.>; 抄送: "Fenger @.**@.>; 主题: Re: [cheng-01037/Self-supervised-Fewshot-Medical-Image-Segmentation] Could not obtain the same prediction results as represented in the paper. (#6)
Hi, can you run CT test successfully? MRI test have no error, but when I test CT, the following problems arise:
— You are receiving this because you commented. Reply to this email directly, view it on GitHub, or unsubscribe.
the reason is that there is a problem with the code in the "Synapse_abdominal_classmap.ipynb" file that processes the SABS data set. The original code :

Please replace this part of the red box code (as shown in the figure below) directly with this part of "class_slice_index_gen.ipynb" in CHAOST2 dir, and then re-run to generate superpixel label for CT images.

The details is: the number of slices read by "nio.read_nii_bysitk" is in the first dimension, that is "n_slice=shape[0]". According to this understanding, the label can be processed smoothly. Good Luck!
@Fenganzhuliang, @DpDark @Liurchen,
I have been able to run the training shell script with no errors, however, I am unable to test. Any time I run the test shell script I get an error that points to the absence of a config file. Please can you assist me on how to solve this. I have attached the details below.
=================================== test_vfold0 sacred.utils.NamedConfigNotFoundError: Named config not found: "feed". Available config values are: ()

How to solve the "config not found"
@Fenganzhuliang, @DpDark @Liurchen, I have been able to run the training shell script with no errors, however, I am unable to test. Any time I run the test shell script I get an error that points to the absence of a config file. Please can you assist me on how to solve this. I have attached the details below.
=================================== test_vfold0 sacred.utils.NamedConfigNotFoundError: Named config not found: "feed". Available config values are: ()
Maybe you need set the right "RELOAD_PATH" in test.sh, after all you can only find the words "feed" in it.
Hello @DpDark,
Thanks for the response. Yet I am getting another error "IndexError". And to be more specific I am trying to run the CT test. The MRI test has run with no issues at all. I have also replaced the part @Fenganzhuliang mentioned but the problem won't fix. Below are the screen shots.


Is there something I am doing wrong?
Hello @DpDark, Thanks for the response. Yet I am getting another error "IndexError". And to be more specific I am trying to run the CT test. The MRI test has run with no issues at all. I have also replaced the part @Fenganzhuliang mentioned but the problem won't fix. Below are the screen shots.
Is there something I am doing wrong?
This is the modified code:

@DpDark, Thanks for the response. I have done the changes and done everything from scratch yet the error still persists. Please check the code below to see from the two modules and see if I am doing something wrong. get_support function from "ManualAnnoDatasetv2.py"
`
def get_support(self, curr_class: int, class_idx: list, scan_idx: list, npart: int):
"""
getting (probably multi-shot) support set for evaluation
sample from 50% (1shot) or 20 35 50 65 80 (5shot)
Args:
curr_cls: current class to segment, starts from 1
class_idx: a list of all foreground class in nways, starts from 1
npart: how may chunks used to split the support
scan_idx: a list, indicating the current **i_th** (note this is idx not pid) training scan
being served as support, in self.pid_curr_load
"""
assert npart % 2 == 1
assert curr_class != 0; assert 0 not in class_idx
assert not self.is_train
self.potential_support_sid = [self.pid_curr_load[ii] for ii in scan_idx ]
print(f'###### Using {len(scan_idx)} shot evaluation!')
if npart == 1:
pcts = [0.5]
else:
half_part = 1 / (npart * 2)
part_interval = (1.0 - 1.0 / npart) / (npart - 1)
pcts = [ half_part + part_interval * ii for ii in range(npart) ]
print(f'###### Parts percentage: {pcts} ######')
out_buffer = [] # [{scanid, img, lb}]
for _part in range(npart):
concat_buffer = [] # for each fold do a concat in image and mask in batch dimension`
for scan_order in scan_idx:
_scan_id = self.pid_curr_load[ scan_order ]
print(f'Using scan {_scan_id} as support!')
# for _pc in pcts:
_zlist = self.tp1_cls_map[self.label_name[curr_class]][_scan_id] # list of indices
_zid = _zlist[int(pcts[_part] * len(_zlist))]
_glb_idx = self.scan_z_idx[_scan_id][_zid]
# almost copy-paste __getitem__ but no augmentation
curr_dict = self.actual_dataset[_glb_idx]
img = curr_dict['img']
lb = curr_dict['lb']
img = np.float32(img)
lb = np.float32(lb).squeeze(-1) # NOTE: to be suitable for the PANet structure
img = torch.from_numpy( np.transpose(img, (2, 0, 1)) )
lb = torch.from_numpy( lb )
if self.tile_z_dim:
img = img.repeat( [ self.tile_z_dim, 1, 1] )
assert img.ndimension() == 3, f'actual dim {img.ndimension()}'
is_start = curr_dict["is_start"]
is_end = curr_dict["is_end"]
nframe = np.int32(curr_dict["nframe"])
scan_id = curr_dict["scan_id"]
z_id = curr_dict["z_id"]
sample = {"image": img,
"label":lb,
"is_start": is_start,
"inst": None,
"scribble": None,
"is_end": is_end,
"nframe": nframe,
"scan_id": scan_id,
"z_id": z_id
}
concat_buffer.append(sample)
out_buffer.append({
"image": torch.stack([itm["image"] for itm in concat_buffer], dim = 0),
"label": torch.stack([itm["label"] for itm in concat_buffer], dim = 0),
})
# do the concat, and add to output_buffer
# post-processing, including keeping the foreground and suppressing background.
support_images = []
support_mask = []
support_class = []
for itm in out_buffer:
support_images.append(itm["image"])
support_class.append(curr_class)
support_mask.append( self.getMaskMedImg( itm["label"], curr_class, class_idx ))
return {'class_ids': [support_class],
'support_images': [support_images], #
'support_mask': [support_mask],
}`
error section of the code from the "Validation.py" file ` with torch.no_grad(): save_pred_buffer = {} # indexed by class
for curr_lb in test_labels:
te_dataset.set_curr_cls(curr_lb)
support_batched = te_parent.get_support(curr_class = curr_lb, class_idx = [curr_lb], scan_idx = _config["support_idx"], npart=_config['task']['npart'])
# way(1 for now) x part x shot x 3 x H x W] #
support_images = [[shot.cuda() for shot in way]
for way in support_batched['support_images']] # way x part x [shot x C x H x W]
suffix = 'mask'
support_fg_mask = [[shot[f'fg_{suffix}'].float().cuda() for shot in way]
for way in support_batched['support_mask']]
support_bg_mask = [[shot[f'bg_{suffix}'].float().cuda() for shot in way]
for way in support_batched['support_mask']]
curr_scan_count = -1 # counting for current scan
_lb_buffer = {} # indexed by scan
last_qpart = 0 # used as indicator for adding result to buffer
for sample_batched in testloader:
_scan_id = sample_batched["scan_id"][0] # we assume batch size for query is 1
if _scan_id in te_parent.potential_support_sid: # skip the support scan, don't include that to query
continue
if sample_batched["is_start"]:
ii = 0
curr_scan_count += 1
_scan_id = sample_batched["scan_id"][0]
outsize = te_dataset.dataset.info_by_scan[_scan_id]["array_size"]
outsize = (256, 256, outsize[0]) # original image read by itk: Z, H, W, in prediction we use H, W, Z
_pred = np.zeros( outsize )
_pred.fill(np.nan)
q_part = sample_batched["part_assign"] # the chunck of query, for assignment with support
query_images = [sample_batched['image'].cuda()]
query_labels = torch.cat([ sample_batched['label'].cuda()], dim=0)
# [way, [part, [shot x C x H x W]]] ->`
Also in the image below is the change that has been done.

@DpDark, Thanks for the response. I have done the changes and done everything from scratch yet the error still persists. Please check the code below to see from the two modules and see if I am doing something wrong. get_support function from "ManualAnnoDatasetv2.py"
`
def get_support(self, curr_class: int, class_idx: list, scan_idx: list, npart: int): """ getting (probably multi-shot) support set for evaluation sample from 50% (1shot) or 20 35 50 65 80 (5shot) Args: curr_cls: current class to segment, starts from 1 class_idx: a list of all foreground class in nways, starts from 1 npart: how may chunks used to split the support scan_idx: a list, indicating the current **i_th** (note this is idx not pid) training scan being served as support, in self.pid_curr_load """ assert npart % 2 == 1 assert curr_class != 0; assert 0 not in class_idx assert not self.is_train self.potential_support_sid = [self.pid_curr_load[ii] for ii in scan_idx ] print(f'###### Using {len(scan_idx)} shot evaluation!') if npart == 1: pcts = [0.5] else: half_part = 1 / (npart * 2) part_interval = (1.0 - 1.0 / npart) / (npart - 1) pcts = [ half_part + part_interval * ii for ii in range(npart) ] print(f'###### Parts percentage: {pcts} ######') out_buffer = [] # [{scanid, img, lb}] for _part in range(npart): concat_buffer = [] # for each fold do a concat in image and mask in batch dimension` for scan_order in scan_idx: _scan_id = self.pid_curr_load[ scan_order ] print(f'Using scan {_scan_id} as support!') # for _pc in pcts: _zlist = self.tp1_cls_map[self.label_name[curr_class]][_scan_id] # list of indices _zid = _zlist[int(pcts[_part] * len(_zlist))] _glb_idx = self.scan_z_idx[_scan_id][_zid] # almost copy-paste __getitem__ but no augmentation curr_dict = self.actual_dataset[_glb_idx] img = curr_dict['img'] lb = curr_dict['lb'] img = np.float32(img) lb = np.float32(lb).squeeze(-1) # NOTE: to be suitable for the PANet structure img = torch.from_numpy( np.transpose(img, (2, 0, 1)) ) lb = torch.from_numpy( lb ) if self.tile_z_dim: img = img.repeat( [ self.tile_z_dim, 1, 1] ) assert img.ndimension() == 3, f'actual dim {img.ndimension()}' is_start = curr_dict["is_start"] is_end = curr_dict["is_end"] nframe = np.int32(curr_dict["nframe"]) scan_id = curr_dict["scan_id"] z_id = curr_dict["z_id"] sample = {"image": img, "label":lb, "is_start": is_start, "inst": None, "scribble": None, "is_end": is_end, "nframe": nframe, "scan_id": scan_id, "z_id": z_id } concat_buffer.append(sample) out_buffer.append({ "image": torch.stack([itm["image"] for itm in concat_buffer], dim = 0), "label": torch.stack([itm["label"] for itm in concat_buffer], dim = 0), }) # do the concat, and add to output_buffer # post-processing, including keeping the foreground and suppressing background. support_images = [] support_mask = [] support_class = [] for itm in out_buffer: support_images.append(itm["image"]) support_class.append(curr_class) support_mask.append( self.getMaskMedImg( itm["label"], curr_class, class_idx )) return {'class_ids': [support_class], 'support_images': [support_images], # 'support_mask': [support_mask], }`
error section of the code from the "Validation.py" file ` with torch.no_grad(): save_pred_buffer = {} # indexed by class
for curr_lb in test_labels: te_dataset.set_curr_cls(curr_lb) support_batched = te_parent.get_support(curr_class = curr_lb, class_idx = [curr_lb], scan_idx = _config["support_idx"], npart=_config['task']['npart']) # way(1 for now) x part x shot x 3 x H x W] # support_images = [[shot.cuda() for shot in way] for way in support_batched['support_images']] # way x part x [shot x C x H x W] suffix = 'mask' support_fg_mask = [[shot[f'fg_{suffix}'].float().cuda() for shot in way] for way in support_batched['support_mask']] support_bg_mask = [[shot[f'bg_{suffix}'].float().cuda() for shot in way] for way in support_batched['support_mask']] curr_scan_count = -1 # counting for current scan _lb_buffer = {} # indexed by scan last_qpart = 0 # used as indicator for adding result to buffer for sample_batched in testloader: _scan_id = sample_batched["scan_id"][0] # we assume batch size for query is 1 if _scan_id in te_parent.potential_support_sid: # skip the support scan, don't include that to query continue if sample_batched["is_start"]: ii = 0 curr_scan_count += 1 _scan_id = sample_batched["scan_id"][0] outsize = te_dataset.dataset.info_by_scan[_scan_id]["array_size"] outsize = (256, 256, outsize[0]) # original image read by itk: Z, H, W, in prediction we use H, W, Z _pred = np.zeros( outsize ) _pred.fill(np.nan) q_part = sample_batched["part_assign"] # the chunck of query, for assignment with support query_images = [sample_batched['image'].cuda()] query_labels = torch.cat([ sample_batched['label'].cuda()], dim=0) # [way, [part, [shot x C x H x W]]] ->`
Also in the image below is the change that has been done.
I think you can restart the whole project code to avoid some careless errors. In fact, just modify the above code, you can run the test with no wrong, so if you made any other modification in this project code, you need to check it again.
As mentioned in the paper, the ABD-30 dataset contains 30 CT scans, but your download only has 20 images with labels? So why?
Hi CHENG, thanks for sharing the code. I am running the code on the Combined Healthy Abdominal Organ Segmentation dataset. While after training, the result of the evaluation is quite different from that represented in the paper. I am wondering whether do I have some misoperation. Here my steps are shown below: At first, I downloaded datasets from https://chaos.grand-challenge.org/Download/. And I got datasets like this:
Then as mentioned in the README.md file, I put the MR folder into
Self-supervised-Fewshot-Medical-Image-Segmentation/data/CHAOST2. In the data pre-processing, first I run./data/CHAOST2/dcm_img_to_nii.shand./data/CHAOST2/png_gth_to_nii.ipynpto convert dicom images to nifti files. After that, I got a folder namedniis/T2SPIR, it consists of images in.nii.gzformat.Then I run
./data/CHAOST2/image_normalize.ipynbto do the pre-processing of the download images, and got normalized image folderchaos_MR_T2_normalized. Next I run./data/CHAOST2/class_slice_index_gen.ipynbto build class-slice indexing for setting up experiments. Then to generate pseudo label, run./data/pseudolabel_gen.ipynb, after all this done, the folder I got is shown below:Then I started to train the model by running
./examples/train_ssl_abdominal_mri.shand here my config file is shown below:""" Experiment configuration file Extended from config file from original PANet Repository """ import os import re import glob import itertools import sacred from sacred import Experiment from sacred.observers import FileStorageObserver from sacred.utils import apply_backspaces_and_linefeeds from platform import node from datetime import datetime sacred.SETTINGS['CONFIG']['READ_ONLY_CONFIG'] = False sacred.SETTINGS.CAPTURE_MODE = 'no' ex = Experiment('mySSL') ex.captured_out_filter = apply_backspaces_and_linefeeds source_folders = ['.', './dataloaders', './models', './util'] sources_to_save = list(itertools.chain.from_iterable( [glob.glob(f'{folder}/*.py') for folder in source_folders])) for source_file in sources_to_save: ex.add_source_file(source_file) @ex.config def cfg(): """Default configurations""" seed = 1234 gpu_id = 0 mode = 'train' # for now only allows 'train' num_workers = 4 # 0 for debugging. dataset = 'CHAOST2_Superpix' # i.e. abdominal MRI use_coco_init = True # initialize backbone with MS_COCO initialization. Anyway coco does not contain medical images ### Training n_steps = 100100 batch_size = 1 lr_milestones = [ (ii + 1) * 1000 for ii in range(n_steps // 1000 - 1)] lr_step_gamma = 0.95 ignore_label = 255 print_interval = 100 save_snapshot_every = 25000 max_iters_per_load = 1000 # epoch size, interval for reloading the dataset scan_per_load = -1 # numbers of 3d scans per load for saving memory. If -1, load the entire dataset to the memory which_aug = 'sabs_aug' # standard data augmentation with intensity and geometric transforms input_size = (256, 256) min_fg_data='100' # when training with manual annotations, indicating number of foreground pixels in a single class single slice. This empirically stablizes the training process label_sets = 0 # which group of labels taking as training (the rest are for testing) exclude_cls_list = [2, 3] # testing classes to be excluded in training. Set to [] if testing under setting 1 usealign = True # see vanilla PANet use_wce = True ### Validation z_margin = 0 eval_fold = 0 # which fold for 5 fold cross validation support_idx=[-1] # indicating which scan is used as support in testing. val_wsize=2 # L_H, L_W in testing n_sup_part = 3 # number of chuncks in testing # Network modelname = 'dlfcn_res101' # resnet 101 backbone from torchvision fcn-deeplab clsname = None # reload_model_path = None # path for reloading a trained model (overrides ms-coco initialization) proto_grid_size = 8 # L_H, L_W = (32, 32) / 8 = (4, 4) in training feature_hw = [32, 32] # feature map size, should couple this with backbone in future # SSL superpix_scale = 'MIDDLE' #MIDDLE/ LARGE model = { 'align': usealign, 'use_coco_init': use_coco_init, 'which_model': modelname, 'cls_name': clsname, 'proto_grid_size' : proto_grid_size, 'feature_hw': feature_hw, 'reload_model_path': reload_model_path } task = { 'n_ways': 1, 'n_shots': 1, 'n_queries': 1, 'npart': n_sup_part } optim_type = 'sgd' optim = { 'lr': 1e-3, 'momentum': 0.9, 'weight_decay': 0.0005, } exp_prefix = '' exp_str = '_'.join( [exp_prefix] + [dataset,] + [f'sets_{label_sets}_{task["n_shots"]}shot']) path = { 'log_dir': './runs', 'SABS':{'data_dir': "/mnt/c/ubunturoot/Self-supervised-Fewshot-Medical-Image-Segmentation/data/SABS/sabs_CT_normalized" }, 'C0':{'data_dir': "feed your dataset path here" }, 'CHAOST2':{'data_dir': "/home/rc/Documents/Self-supervised-Fewshot-Medical-Image-Segmentation/data/CHAOST2/chaos_MR_T2_normalized/" }, 'SABS_Superpix':{'data_dir': "/mnt/c/ubunturoot/Self-supervised-Fewshot-Medical-Image-Segmentation/data/SABS/sabs_CT_normalized"}, 'C0_Superpix':{'data_dir': "feed your dataset path here"}, 'CHAOST2_Superpix':{'data_dir': "/home/rc/Documents/Self-supervised-Fewshot-Medical-Image-Segmentation/data/CHAOST2/chaos_MR_T2_normalized/"}, } @ex.config_hook def add_observer(config, command_name, logger): """A hook fucntion to add observer""" exp_name = f'{ex.path}_{config["exp_str"]}' observer = FileStorageObserver.create(os.path.join(config['path']['log_dir'], exp_name)) ex.observers.append(observer) return configAfter training, I have got some snapshot of the model:
Then I run
./examples/test_ssl_abdominal_mri.shto test the model, and below is my result:As is shown in the picture, the dice score is about 0.8, while in the paper, it shoule be around 80, it's quite different. So I wonder if there are some mistakes during my experiment? And how could I fix it and get ideal results? I would deeply appreciate your reply.
Hello, the data in the paper is a percentage, which means that 0.8 is represented by 80%, and the "%" is omitted in the table. Your results are no problem.
Hi, I'm trying to do some changes on this model. But I have a question about the CT dataset. Could we exchange our email or something?
And my eamil is kangnniexiao@163.com
@DpDark, Thanks for the response. I have done the changes and done everything from scratch yet the error still persists. Please check the code below to see from the two modules and see if I am doing something wrong. get_support function from "ManualAnnoDatasetv2.py"
`
def get_support(self, curr_class: int, class_idx: list, scan_idx: list, npart: int): """ getting (probably multi-shot) support set for evaluation sample from 50% (1shot) or 20 35 50 65 80 (5shot) Args: curr_cls: current class to segment, starts from 1 class_idx: a list of all foreground class in nways, starts from 1 npart: how may chunks used to split the support scan_idx: a list, indicating the current **i_th** (note this is idx not pid) training scan being served as support, in self.pid_curr_load """ assert npart % 2 == 1 assert curr_class != 0; assert 0 not in class_idx assert not self.is_train self.potential_support_sid = [self.pid_curr_load[ii] for ii in scan_idx ] print(f'###### Using {len(scan_idx)} shot evaluation!') if npart == 1: pcts = [0.5] else: half_part = 1 / (npart * 2) part_interval = (1.0 - 1.0 / npart) / (npart - 1) pcts = [ half_part + part_interval * ii for ii in range(npart) ] print(f'###### Parts percentage: {pcts} ######') out_buffer = [] # [{scanid, img, lb}] for _part in range(npart): concat_buffer = [] # for each fold do a concat in image and mask in batch dimension` for scan_order in scan_idx: _scan_id = self.pid_curr_load[ scan_order ] print(f'Using scan {_scan_id} as support!') # for _pc in pcts: _zlist = self.tp1_cls_map[self.label_name[curr_class]][_scan_id] # list of indices _zid = _zlist[int(pcts[_part] * len(_zlist))] _glb_idx = self.scan_z_idx[_scan_id][_zid] # almost copy-paste __getitem__ but no augmentation curr_dict = self.actual_dataset[_glb_idx] img = curr_dict['img'] lb = curr_dict['lb'] img = np.float32(img) lb = np.float32(lb).squeeze(-1) # NOTE: to be suitable for the PANet structure img = torch.from_numpy( np.transpose(img, (2, 0, 1)) ) lb = torch.from_numpy( lb ) if self.tile_z_dim: img = img.repeat( [ self.tile_z_dim, 1, 1] ) assert img.ndimension() == 3, f'actual dim {img.ndimension()}' is_start = curr_dict["is_start"] is_end = curr_dict["is_end"] nframe = np.int32(curr_dict["nframe"]) scan_id = curr_dict["scan_id"] z_id = curr_dict["z_id"] sample = {"image": img, "label":lb, "is_start": is_start, "inst": None, "scribble": None, "is_end": is_end, "nframe": nframe, "scan_id": scan_id, "z_id": z_id } concat_buffer.append(sample) out_buffer.append({ "image": torch.stack([itm["image"] for itm in concat_buffer], dim = 0), "label": torch.stack([itm["label"] for itm in concat_buffer], dim = 0), }) # do the concat, and add to output_buffer # post-processing, including keeping the foreground and suppressing background. support_images = [] support_mask = [] support_class = [] for itm in out_buffer: support_images.append(itm["image"]) support_class.append(curr_class) support_mask.append( self.getMaskMedImg( itm["label"], curr_class, class_idx )) return {'class_ids': [support_class], 'support_images': [support_images], # 'support_mask': [support_mask], }`
error section of the code from the "Validation.py" file ` with torch.no_grad(): save_pred_buffer = {} # indexed by class
for curr_lb in test_labels: te_dataset.set_curr_cls(curr_lb) support_batched = te_parent.get_support(curr_class = curr_lb, class_idx = [curr_lb], scan_idx = _config["support_idx"], npart=_config['task']['npart']) # way(1 for now) x part x shot x 3 x H x W] # support_images = [[shot.cuda() for shot in way] for way in support_batched['support_images']] # way x part x [shot x C x H x W] suffix = 'mask' support_fg_mask = [[shot[f'fg_{suffix}'].float().cuda() for shot in way] for way in support_batched['support_mask']] support_bg_mask = [[shot[f'bg_{suffix}'].float().cuda() for shot in way] for way in support_batched['support_mask']] curr_scan_count = -1 # counting for current scan _lb_buffer = {} # indexed by scan last_qpart = 0 # used as indicator for adding result to buffer for sample_batched in testloader: _scan_id = sample_batched["scan_id"][0] # we assume batch size for query is 1 if _scan_id in te_parent.potential_support_sid: # skip the support scan, don't include that to query continue if sample_batched["is_start"]: ii = 0 curr_scan_count += 1 _scan_id = sample_batched["scan_id"][0] outsize = te_dataset.dataset.info_by_scan[_scan_id]["array_size"] outsize = (256, 256, outsize[0]) # original image read by itk: Z, H, W, in prediction we use H, W, Z _pred = np.zeros( outsize ) _pred.fill(np.nan) q_part = sample_batched["part_assign"] # the chunck of query, for assignment with support query_images = [sample_batched['image'].cuda()] query_labels = torch.cat([ sample_batched['label'].cuda()], dim=0) # [way, [part, [shot x C x H x W]]] ->`
Also in the image below is the change that has been done.
@DpDark, Thanks for the response. I have done the changes and done everything from scratch yet the error still persists. Please check the code below to see from the two modules and see if I am doing something wrong. get_support function from "ManualAnnoDatasetv2.py"
`
def get_support(self, curr_class: int, class_idx: list, scan_idx: list, npart: int): """ getting (probably multi-shot) support set for evaluation sample from 50% (1shot) or 20 35 50 65 80 (5shot) Args: curr_cls: current class to segment, starts from 1 class_idx: a list of all foreground class in nways, starts from 1 npart: how may chunks used to split the support scan_idx: a list, indicating the current **i_th** (note this is idx not pid) training scan being served as support, in self.pid_curr_load """ assert npart % 2 == 1 assert curr_class != 0; assert 0 not in class_idx assert not self.is_train self.potential_support_sid = [self.pid_curr_load[ii] for ii in scan_idx ] print(f'###### Using {len(scan_idx)} shot evaluation!') if npart == 1: pcts = [0.5] else: half_part = 1 / (npart * 2) part_interval = (1.0 - 1.0 / npart) / (npart - 1) pcts = [ half_part + part_interval * ii for ii in range(npart) ] print(f'###### Parts percentage: {pcts} ######') out_buffer = [] # [{scanid, img, lb}] for _part in range(npart): concat_buffer = [] # for each fold do a concat in image and mask in batch dimension` for scan_order in scan_idx: _scan_id = self.pid_curr_load[ scan_order ] print(f'Using scan {_scan_id} as support!') # for _pc in pcts: _zlist = self.tp1_cls_map[self.label_name[curr_class]][_scan_id] # list of indices _zid = _zlist[int(pcts[_part] * len(_zlist))] _glb_idx = self.scan_z_idx[_scan_id][_zid] # almost copy-paste __getitem__ but no augmentation curr_dict = self.actual_dataset[_glb_idx] img = curr_dict['img'] lb = curr_dict['lb'] img = np.float32(img) lb = np.float32(lb).squeeze(-1) # NOTE: to be suitable for the PANet structure img = torch.from_numpy( np.transpose(img, (2, 0, 1)) ) lb = torch.from_numpy( lb ) if self.tile_z_dim: img = img.repeat( [ self.tile_z_dim, 1, 1] ) assert img.ndimension() == 3, f'actual dim {img.ndimension()}' is_start = curr_dict["is_start"] is_end = curr_dict["is_end"] nframe = np.int32(curr_dict["nframe"]) scan_id = curr_dict["scan_id"] z_id = curr_dict["z_id"] sample = {"image": img, "label":lb, "is_start": is_start, "inst": None, "scribble": None, "is_end": is_end, "nframe": nframe, "scan_id": scan_id, "z_id": z_id } concat_buffer.append(sample) out_buffer.append({ "image": torch.stack([itm["image"] for itm in concat_buffer], dim = 0), "label": torch.stack([itm["label"] for itm in concat_buffer], dim = 0), }) # do the concat, and add to output_buffer # post-processing, including keeping the foreground and suppressing background. support_images = [] support_mask = [] support_class = [] for itm in out_buffer: support_images.append(itm["image"]) support_class.append(curr_class) support_mask.append( self.getMaskMedImg( itm["label"], curr_class, class_idx )) return {'class_ids': [support_class], 'support_images': [support_images], # 'support_mask': [support_mask], }`
error section of the code from the "Validation.py" file ` with torch.no_grad(): save_pred_buffer = {} # indexed by class
for curr_lb in test_labels: te_dataset.set_curr_cls(curr_lb) support_batched = te_parent.get_support(curr_class = curr_lb, class_idx = [curr_lb], scan_idx = _config["support_idx"], npart=_config['task']['npart']) # way(1 for now) x part x shot x 3 x H x W] # support_images = [[shot.cuda() for shot in way] for way in support_batched['support_images']] # way x part x [shot x C x H x W] suffix = 'mask' support_fg_mask = [[shot[f'fg_{suffix}'].float().cuda() for shot in way] for way in support_batched['support_mask']] support_bg_mask = [[shot[f'bg_{suffix}'].float().cuda() for shot in way] for way in support_batched['support_mask']] curr_scan_count = -1 # counting for current scan _lb_buffer = {} # indexed by scan last_qpart = 0 # used as indicator for adding result to buffer for sample_batched in testloader: _scan_id = sample_batched["scan_id"][0] # we assume batch size for query is 1 if _scan_id in te_parent.potential_support_sid: # skip the support scan, don't include that to query continue if sample_batched["is_start"]: ii = 0 curr_scan_count += 1 _scan_id = sample_batched["scan_id"][0] outsize = te_dataset.dataset.info_by_scan[_scan_id]["array_size"] outsize = (256, 256, outsize[0]) # original image read by itk: Z, H, W, in prediction we use H, W, Z _pred = np.zeros( outsize ) _pred.fill(np.nan) q_part = sample_batched["part_assign"] # the chunck of query, for assignment with support query_images = [sample_batched['image'].cuda()] query_labels = torch.cat([ sample_batched['label'].cuda()], dim=0) # [way, [part, [shot x C x H x W]]] ->`
Also in the image below is the change that has been done.
Hi Ashong-Nartey, when i tried to run test_ssl_abdominal_ct.sh ,the same problem occurred. Have you solved it?Can you give me some advice,thanks.
@Nothingloser , I was not able to run the test_ssl_abdominal_ct.sh. The train_ssl_abdominal_ct.sh however could run successfully, likewise, the CHAOS training and test was successful. please contact @Liurchen and @DpDark.
How to solve the "config not found"
@Fenganzhuliang, @DpDark @Liurchen, I have been able to run the training shell script with no errors, however, I am unable to test. Any time I run the test shell script I get an error that points to the absence of a config file. Please can you assist me on how to solve this. I have attached the details below.
=================================== test_vfold0 sacred.utils.NamedConfigNotFoundError: Named config not found: "feed". Available config values are: ()
@Ashong-Nartey i have problem when i run the test_ssl_abdominal_mri .please can you assist me on how to solve this

I I have set path for RELOAD_PATH. but I still got this problem
@DpDark, Thanks for the response. I have done the changes and done everything from scratch yet the error still persists. Please check the code below to see from the two modules and see if I am doing something wrong. get_support function from "ManualAnnoDatasetv2.py" `
def get_support(self, curr_class: int, class_idx: list, scan_idx: list, npart: int): """ getting (probably multi-shot) support set for evaluation sample from 50% (1shot) or 20 35 50 65 80 (5shot) Args: curr_cls: current class to segment, starts from 1 class_idx: a list of all foreground class in nways, starts from 1 npart: how may chunks used to split the support scan_idx: a list, indicating the current **i_th** (note this is idx not pid) training scan being served as support, in self.pid_curr_load """ assert npart % 2 == 1 assert curr_class != 0; assert 0 not in class_idx assert not self.is_train self.potential_support_sid = [self.pid_curr_load[ii] for ii in scan_idx ] print(f'###### Using {len(scan_idx)} shot evaluation!') if npart == 1: pcts = [0.5] else: half_part = 1 / (npart * 2) part_interval = (1.0 - 1.0 / npart) / (npart - 1) pcts = [ half_part + part_interval * ii for ii in range(npart) ] print(f'###### Parts percentage: {pcts} ######') out_buffer = [] # [{scanid, img, lb}] for _part in range(npart): concat_buffer = [] # for each fold do a concat in image and mask in batch dimension` for scan_order in scan_idx: _scan_id = self.pid_curr_load[ scan_order ] print(f'Using scan {_scan_id} as support!') # for _pc in pcts: _zlist = self.tp1_cls_map[self.label_name[curr_class]][_scan_id] # list of indices _zid = _zlist[int(pcts[_part] * len(_zlist))] _glb_idx = self.scan_z_idx[_scan_id][_zid] # almost copy-paste __getitem__ but no augmentation curr_dict = self.actual_dataset[_glb_idx] img = curr_dict['img'] lb = curr_dict['lb'] img = np.float32(img) lb = np.float32(lb).squeeze(-1) # NOTE: to be suitable for the PANet structure img = torch.from_numpy( np.transpose(img, (2, 0, 1)) ) lb = torch.from_numpy( lb ) if self.tile_z_dim: img = img.repeat( [ self.tile_z_dim, 1, 1] ) assert img.ndimension() == 3, f'actual dim {img.ndimension()}' is_start = curr_dict["is_start"] is_end = curr_dict["is_end"] nframe = np.int32(curr_dict["nframe"]) scan_id = curr_dict["scan_id"] z_id = curr_dict["z_id"] sample = {"image": img, "label":lb, "is_start": is_start, "inst": None, "scribble": None, "is_end": is_end, "nframe": nframe, "scan_id": scan_id, "z_id": z_id } concat_buffer.append(sample) out_buffer.append({ "image": torch.stack([itm["image"] for itm in concat_buffer], dim = 0), "label": torch.stack([itm["label"] for itm in concat_buffer], dim = 0), }) # do the concat, and add to output_buffer # post-processing, including keeping the foreground and suppressing background. support_images = [] support_mask = [] support_class = [] for itm in out_buffer: support_images.append(itm["image"]) support_class.append(curr_class) support_mask.append( self.getMaskMedImg( itm["label"], curr_class, class_idx )) return {'class_ids': [support_class], 'support_images': [support_images], # 'support_mask': [support_mask], }
**error section of the code from the "Validation.py" file**with torch.no_grad(): save_pred_buffer = {} # indexed by classfor curr_lb in test_labels: te_dataset.set_curr_cls(curr_lb) support_batched = te_parent.get_support(curr_class = curr_lb, class_idx = [curr_lb], scan_idx = _config["support_idx"], npart=_config['task']['npart']) # way(1 for now) x part x shot x 3 x H x W] # support_images = [[shot.cuda() for shot in way] for way in support_batched['support_images']] # way x part x [shot x C x H x W] suffix = 'mask' support_fg_mask = [[shot[f'fg_{suffix}'].float().cuda() for shot in way] for way in support_batched['support_mask']] support_bg_mask = [[shot[f'bg_{suffix}'].float().cuda() for shot in way] for way in support_batched['support_mask']] curr_scan_count = -1 # counting for current scan _lb_buffer = {} # indexed by scan last_qpart = 0 # used as indicator for adding result to buffer for sample_batched in testloader: _scan_id = sample_batched["scan_id"][0] # we assume batch size for query is 1 if _scan_id in te_parent.potential_support_sid: # skip the support scan, don't include that to query continue if sample_batched["is_start"]: ii = 0 curr_scan_count += 1 _scan_id = sample_batched["scan_id"][0] outsize = te_dataset.dataset.info_by_scan[_scan_id]["array_size"] outsize = (256, 256, outsize[0]) # original image read by itk: Z, H, W, in prediction we use H, W, Z _pred = np.zeros( outsize ) _pred.fill(np.nan) q_part = sample_batched["part_assign"] # the chunck of query, for assignment with support query_images = [sample_batched['image'].cuda()] query_labels = torch.cat([ sample_batched['label'].cuda()], dim=0) # [way, [part, [shot x C x H x W]]] ->` Also in the image below is the change that has been done.
@DpDark, Thanks for the response. I have done the changes and done everything from scratch yet the error still persists. Please check the code below to see from the two modules and see if I am doing something wrong. get_support function from "ManualAnnoDatasetv2.py" `
def get_support(self, curr_class: int, class_idx: list, scan_idx: list, npart: int): """ getting (probably multi-shot) support set for evaluation sample from 50% (1shot) or 20 35 50 65 80 (5shot) Args: curr_cls: current class to segment, starts from 1 class_idx: a list of all foreground class in nways, starts from 1 npart: how may chunks used to split the support scan_idx: a list, indicating the current **i_th** (note this is idx not pid) training scan being served as support, in self.pid_curr_load """ assert npart % 2 == 1 assert curr_class != 0; assert 0 not in class_idx assert not self.is_train self.potential_support_sid = [self.pid_curr_load[ii] for ii in scan_idx ] print(f'###### Using {len(scan_idx)} shot evaluation!') if npart == 1: pcts = [0.5] else: half_part = 1 / (npart * 2) part_interval = (1.0 - 1.0 / npart) / (npart - 1) pcts = [ half_part + part_interval * ii for ii in range(npart) ] print(f'###### Parts percentage: {pcts} ######') out_buffer = [] # [{scanid, img, lb}] for _part in range(npart): concat_buffer = [] # for each fold do a concat in image and mask in batch dimension` for scan_order in scan_idx: _scan_id = self.pid_curr_load[ scan_order ] print(f'Using scan {_scan_id} as support!') # for _pc in pcts: _zlist = self.tp1_cls_map[self.label_name[curr_class]][_scan_id] # list of indices _zid = _zlist[int(pcts[_part] * len(_zlist))] _glb_idx = self.scan_z_idx[_scan_id][_zid] # almost copy-paste __getitem__ but no augmentation curr_dict = self.actual_dataset[_glb_idx] img = curr_dict['img'] lb = curr_dict['lb'] img = np.float32(img) lb = np.float32(lb).squeeze(-1) # NOTE: to be suitable for the PANet structure img = torch.from_numpy( np.transpose(img, (2, 0, 1)) ) lb = torch.from_numpy( lb ) if self.tile_z_dim: img = img.repeat( [ self.tile_z_dim, 1, 1] ) assert img.ndimension() == 3, f'actual dim {img.ndimension()}' is_start = curr_dict["is_start"] is_end = curr_dict["is_end"] nframe = np.int32(curr_dict["nframe"]) scan_id = curr_dict["scan_id"] z_id = curr_dict["z_id"] sample = {"image": img, "label":lb, "is_start": is_start, "inst": None, "scribble": None, "is_end": is_end, "nframe": nframe, "scan_id": scan_id, "z_id": z_id } concat_buffer.append(sample) out_buffer.append({ "image": torch.stack([itm["image"] for itm in concat_buffer], dim = 0), "label": torch.stack([itm["label"] for itm in concat_buffer], dim = 0), }) # do the concat, and add to output_buffer # post-processing, including keeping the foreground and suppressing background. support_images = [] support_mask = [] support_class = [] for itm in out_buffer: support_images.append(itm["image"]) support_class.append(curr_class) support_mask.append( self.getMaskMedImg( itm["label"], curr_class, class_idx )) return {'class_ids': [support_class], 'support_images': [support_images], # 'support_mask': [support_mask], }
**error section of the code from the "Validation.py" file**with torch.no_grad(): save_pred_buffer = {} # indexed by classfor curr_lb in test_labels: te_dataset.set_curr_cls(curr_lb) support_batched = te_parent.get_support(curr_class = curr_lb, class_idx = [curr_lb], scan_idx = _config["support_idx"], npart=_config['task']['npart']) # way(1 for now) x part x shot x 3 x H x W] # support_images = [[shot.cuda() for shot in way] for way in support_batched['support_images']] # way x part x [shot x C x H x W] suffix = 'mask' support_fg_mask = [[shot[f'fg_{suffix}'].float().cuda() for shot in way] for way in support_batched['support_mask']] support_bg_mask = [[shot[f'bg_{suffix}'].float().cuda() for shot in way] for way in support_batched['support_mask']] curr_scan_count = -1 # counting for current scan _lb_buffer = {} # indexed by scan last_qpart = 0 # used as indicator for adding result to buffer for sample_batched in testloader: _scan_id = sample_batched["scan_id"][0] # we assume batch size for query is 1 if _scan_id in te_parent.potential_support_sid: # skip the support scan, don't include that to query continue if sample_batched["is_start"]: ii = 0 curr_scan_count += 1 _scan_id = sample_batched["scan_id"][0] outsize = te_dataset.dataset.info_by_scan[_scan_id]["array_size"] outsize = (256, 256, outsize[0]) # original image read by itk: Z, H, W, in prediction we use H, W, Z _pred = np.zeros( outsize ) _pred.fill(np.nan) q_part = sample_batched["part_assign"] # the chunck of query, for assignment with support query_images = [sample_batched['image'].cuda()] query_labels = torch.cat([ sample_batched['label'].cuda()], dim=0) # [way, [part, [shot x C x H x W]]] ->` Also in the image below is the change that has been done.
Hi Ashong-Nartey, when i tried to run test_ssl_abdominal_ct.sh ,the same problem occurred. Have you solved it?Can you give me some advice,thanks.
 Have you experienced this problem when dealing with CT datasets? KID_R and KID_l are empty on my Classmap.json
Have you experienced this problem when dealing with CT datasets? KID_R and KID_l are empty on my Classmap.json
@Goooo1898 Sorry for the late response. However, I couldn't solve that issue. And the authors have since not responded too.
@Cmnotjx Yes, I just checked and realized those classes are also empty in the classmap.json file. I hope the authors of the repository could clarify these issues.
Thanks for the issues and sorry for the late response. Will run a test and fix them.
@Cmnotjx Yes, I just checked and realized those classes are also empty in the classmap.json file. I hope the authors of the repository could clarify these issues.
Hi, I have solved this question because the raw dataset you have used is incorrect. Please participate in the Synapse challenge in order to download the raw datasets. If you use the datasets which name are "averaged-*", that is the wrong datasets. The real raw datasets of the CT image are:

@Ashong-Nartey You need to run this for both CT and MRI. For MRI you also may want to adjust label names and file paths as well according to those in MRI dataset.
Will leave this issue open for a short while ...
@cheng-01037 and @kentanvictor Alright. I will try it right away and give feedback. Thanks.
What has to be replaced in RELOAD_PATH ? Could anyone please clarify this ?
I I have set path for RELOAD_PATH. but I still got this problem
@viethoang303 Could you please let me know the path that has to be set in RELOAD_PATH ?
Thanks.
I I have set path for RELOAD_PATH. but I still got this problem
@viethoang303 Could you please let me know the path that has to be set in RELOAD_PATH ?
Thanks.
It has been resolved.
@Cmnotjx Yes, I just checked and realized those classes are also empty in the classmap.json file. I hope the authors of the repository could clarify these issues.
Hi, I have solved this question because the raw dataset you have used is incorrect. Please participate in the Synapse challenge in order to download the raw datasets. If you use the datasets which name are "averaged-*", that is the wrong datasets. The real raw datasets of the
CT imageare:
hello @kentanvictor would you mind telling me how to find raw data option in that website?i open it up found there is no raw data option[.]
 (https://www.synapse.org/#!Synapse:syn3193805/files/)
(https://www.synapse.org/#!Synapse:syn3193805/files/)
@Cmnotjx Yes, I just checked and realized those classes are also empty in the classmap.json file. I hope the authors of the repository could clarify these issues.
Hi, I have solved this question because the raw dataset you have used is incorrect. Please participate in the Synapse challenge in order to download the raw datasets. If you use the datasets which name are "averaged-*", that is the wrong datasets. The real raw datasets of the
CT imageare:
I have already solved this problem. Thank you for your sharing!
I I have set path for RELOAD_PATH. but I still got this problem
@viethoang303 Could you please let me know the path that has to be set in RELOAD_PATH ? Thanks.
It has been resolved. hello i don‘t know the mean of ‘’reload_path ’,can you share what the reload path is? thank u.
How to solve the "config not found"
@Fenganzhuliang, @DpDark @Liurchen, I have been able to run the training shell script with no errors, however, I am unable to test. Any time I run the test shell script I get an error that points to the absence of a config file. Please can you assist me on how to solve this. I have attached the details below.
=================================== test_vfold0 sacred.utils.NamedConfigNotFoundError: Named config not found: "feed". Available config values are: ()hello,can you tell me what the reload path is?what is your path?i don't know Thank u!
How to solve the "config not found"
@Fenganzhuliang, @DpDark @Liurchen, I have been able to run the training shell script with no errors, however, I am unable to test. Any time I run the test shell script I get an error that points to the absence of a config file. Please can you assist me on how to solve this. I have attached the details below.
=================================== test_vfold0 sacred.utils.NamedConfigNotFoundError: Named config not found: "feed". Available config values are: ()hello,can you tell me what the reload path is?what is your path?i don't know Thank u!
That is the path to your trained model. You need to load your trained model to the script before testing.
I I have set path for RELOAD_PATH. but I still got this problem
@viethoang303 Could you please let me know the path that has to be set in RELOAD_PATH ? Thanks.
It has been resolved. hello i don‘t know the mean of ‘’reload_path ’,can you share what the reload path is? thank u.
Sorry, where can I find the path for this' RELOAD_PATH '? I did not see the weight. xx file in the model I trained under './exp'.
Hi CHENG, thanks for sharing the code. I am running the code on the Combined Healthy Abdominal Organ Segmentation dataset. While after training, the result of the evaluation is quite different from that represented in the paper. I am wondering whether do I have some misoperation. Here my steps are shown below: At first, I downloaded datasets from https://chaos.grand-challenge.org/Download/. And I got datasets like this: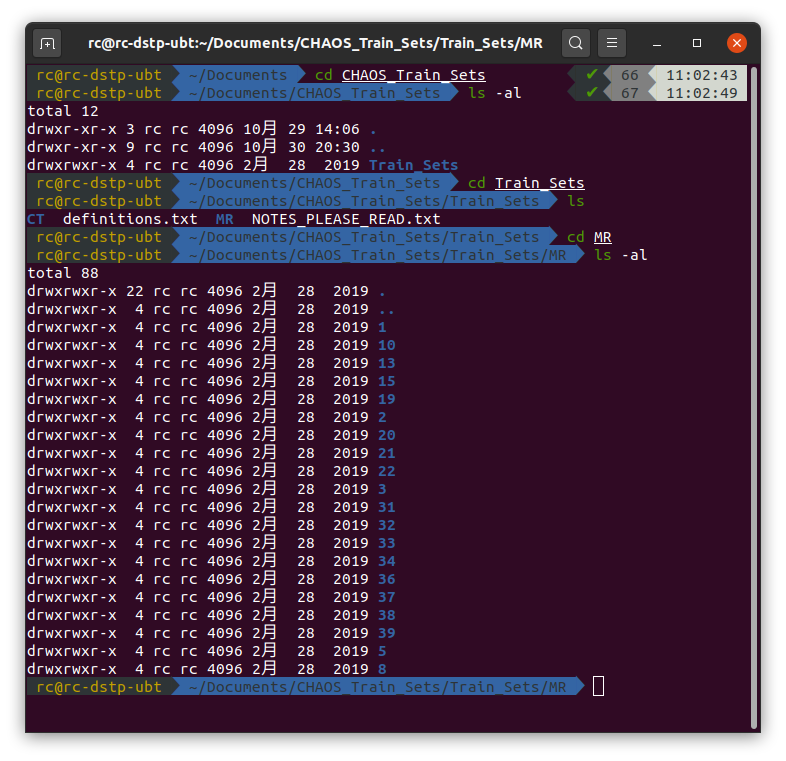 Then as mentioned in the README.md file, I put the MR folder into
Then as mentioned in the README.md file, I put the MR folder into 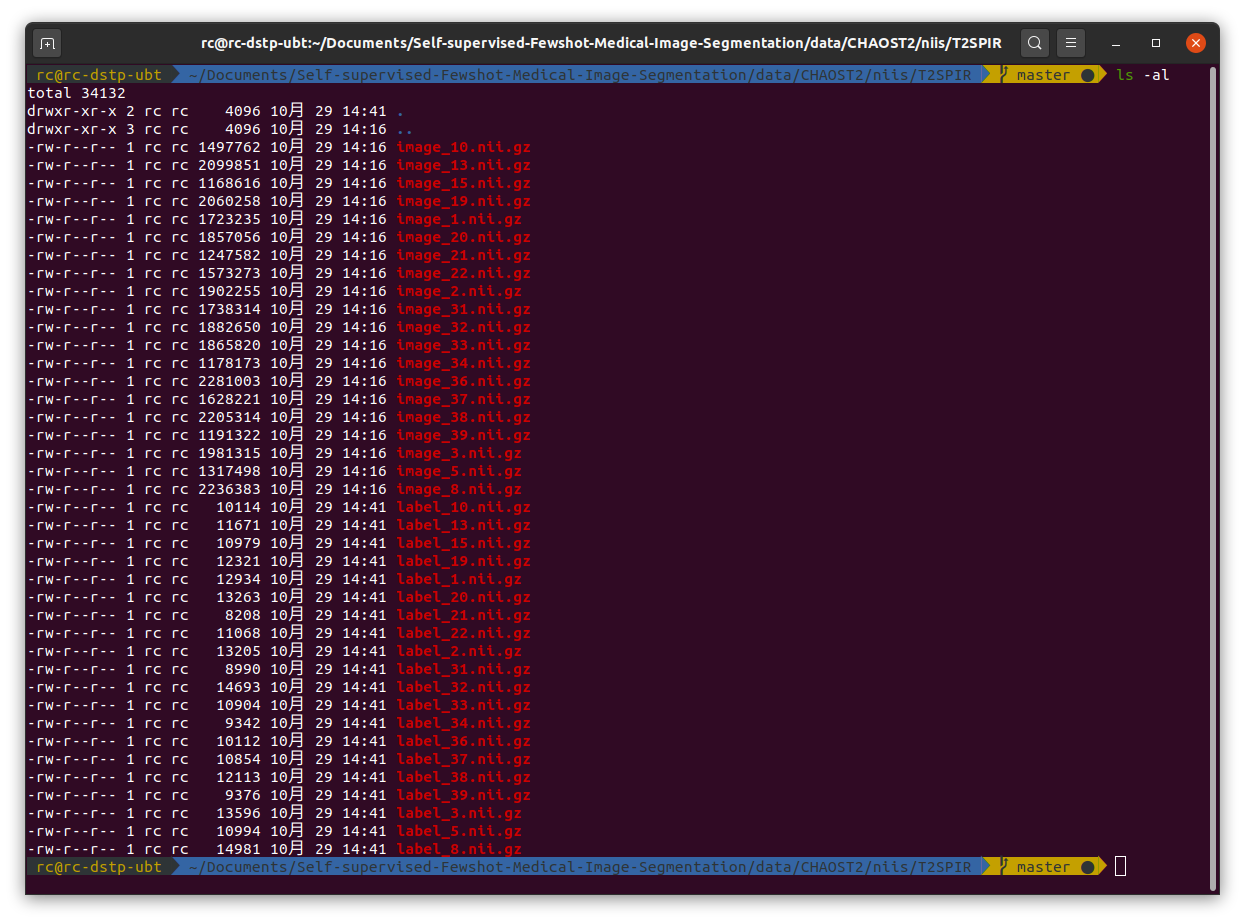 Then I run
Then I run 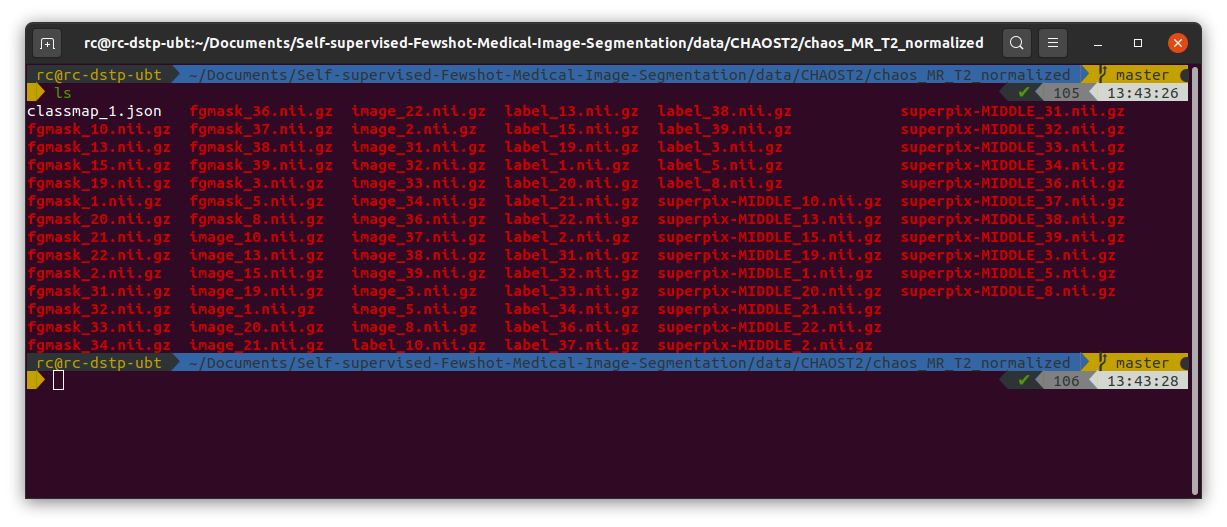 Then I started to train the model by running
Then I started to train the model by running
Self-supervised-Fewshot-Medical-Image-Segmentation/data/CHAOST2. In the data pre-processing, first I run./data/CHAOST2/dcm_img_to_nii.shand./data/CHAOST2/png_gth_to_nii.ipynpto convert dicom images to nifti files. After that, I got a folder namedniis/T2SPIR, it consists of images in.nii.gzformat../data/CHAOST2/image_normalize.ipynbto do the pre-processing of the download images, and got normalized image folderchaos_MR_T2_normalized. Next I run./data/CHAOST2/class_slice_index_gen.ipynbto build class-slice indexing for setting up experiments. Then to generate pseudo label, run./data/pseudolabel_gen.ipynb, after all this done, the folder I got is shown below:./examples/train_ssl_abdominal_mri.shand here my config file is shown below:After training, I have got some snapshot of the model: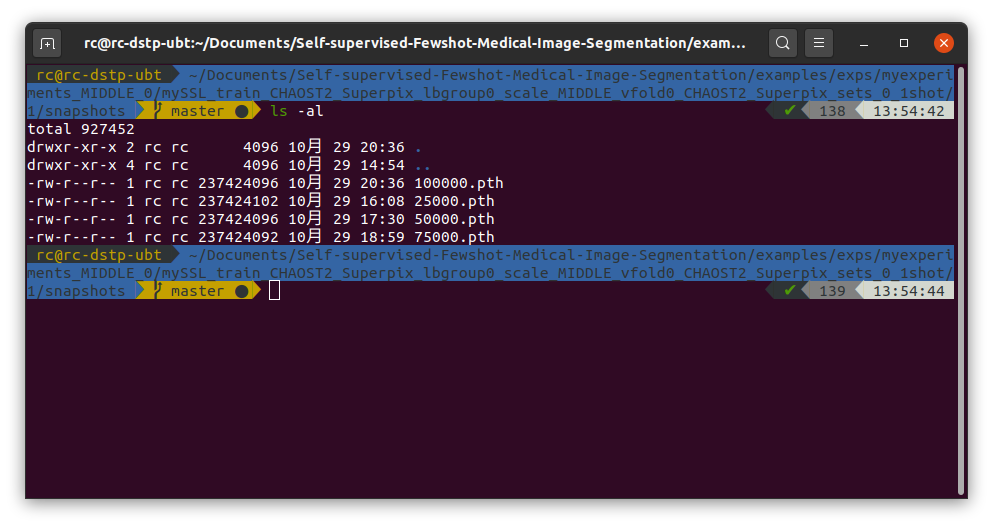 Then I run
Then I run 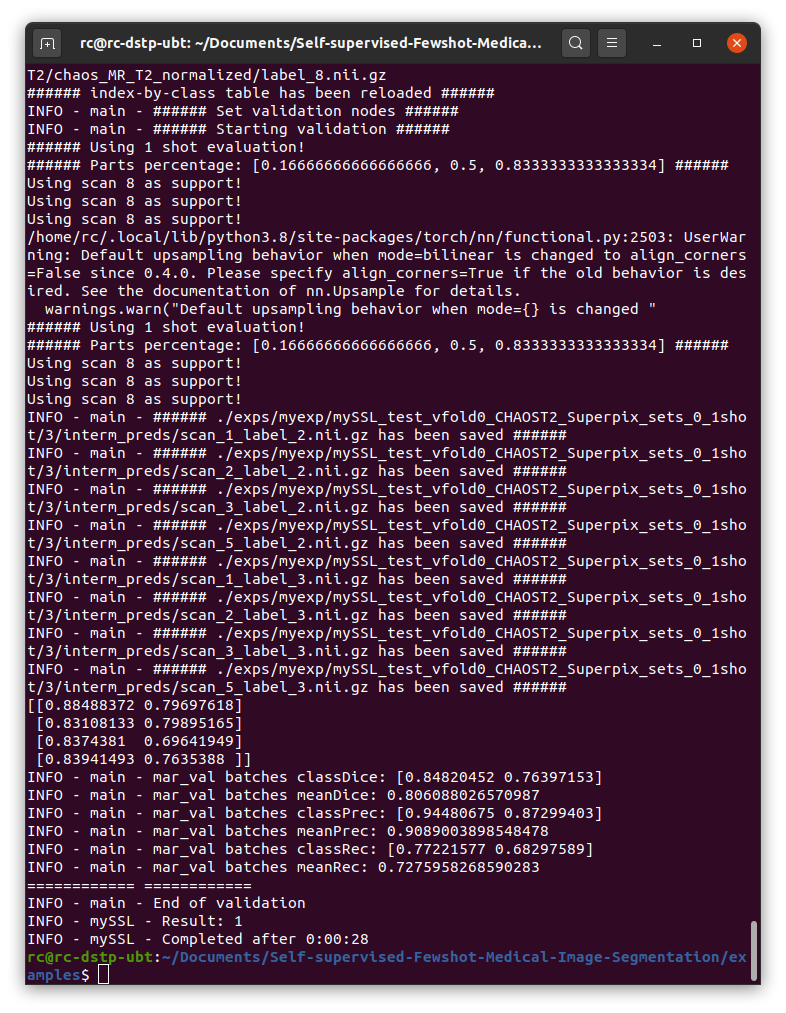 As is shown in the picture, the dice score is about 0.8, while in the paper, it shoule be around 80, it's quite different.
So I wonder if there are some mistakes during my experiment? And how could I fix it and get ideal results?
I would deeply appreciate your reply.
As is shown in the picture, the dice score is about 0.8, while in the paper, it shoule be around 80, it's quite different.
So I wonder if there are some mistakes during my experiment? And how could I fix it and get ideal results?
I would deeply appreciate your reply.
./examples/test_ssl_abdominal_mri.shto test the model, and below is my result: